Evaluating Self-Supervised Learning for Molecular Graph Embeddings
Paper and Code
Jun 16, 2022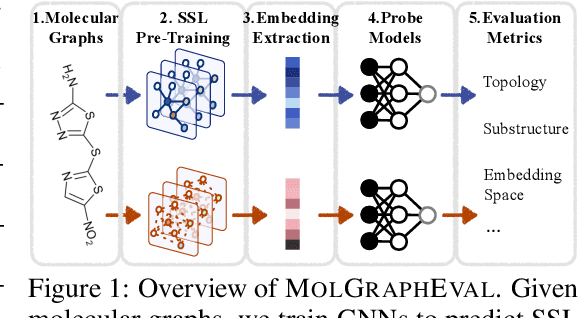
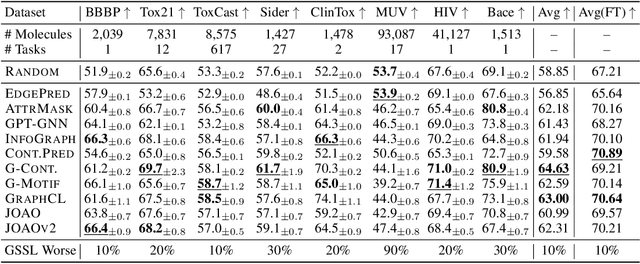
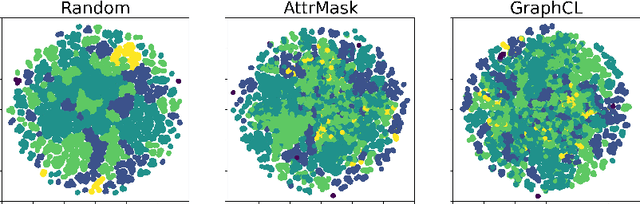
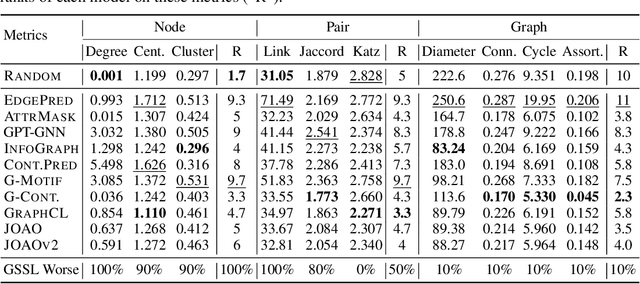
Graph Self-Supervised Learning (GSSL) paves the way for learning graph embeddings without expert annotation, which is particularly impactful for molecular graphs since the number of possible molecules is enormous and labels are expensive to obtain. However, by design, GSSL methods are not trained to perform well on one downstream task but aim for transferability to many, making evaluating them less straightforward. As a step toward obtaining profiles of molecular graph embeddings with diverse and interpretable attributes, we introduce Molecular Graph Representation Evaluation (MolGraphEval), a suite of probe tasks, categorised into (i) topological-, (ii) substructure-, and (iii) embedding space properties. By benchmarking existing GSSL methods on both existing downstream datasets and MolGraphEval, we discover surprising discrepancies between conclusions drawn from existing datasets alone versus more fine-grained probing, suggesting that current evaluation protocols do not provide the whole picture. Our modular, automated end-to-end GSSL pipeline code will be released upon acceptance, including standardised graph loading, experiment management, and embedding evaluation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge