Discovering the Representation Bottleneck of Graph Neural Networks from Multi-order Interactions
Paper and Code
May 21, 2022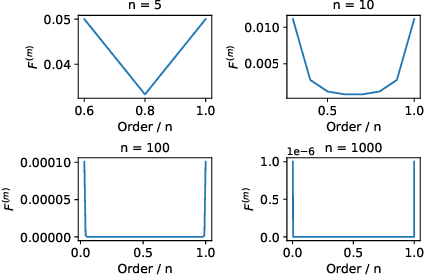
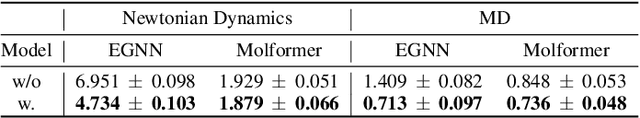
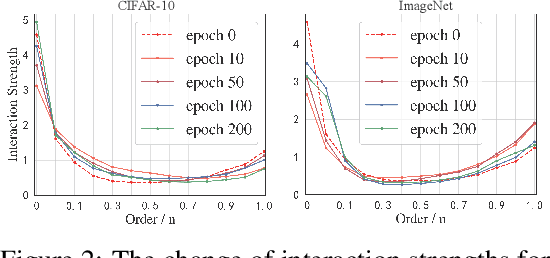
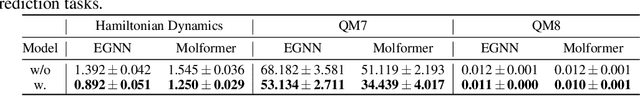
Most graph neural networks (GNNs) rely on the message passing paradigm to propagate node features and build interactions. Recent works point out that different graph learning tasks require different ranges of interactions between nodes. To investigate the underlying mechanism, we explore the capacity of GNNs to capture pairwise interactions between nodes under contexts with different complexities, especially for their graph-level and node-level applications in scientific domains like biochemistry and physics. When formulating pairwise interactions, we study two standard graph construction methods in scientific domains, i.e., K-nearest neighbor (KNN) graphs and fully-connected (FC) graphs. Furthermore, we demonstrate that the inductive bias introduced by KNN-graphs and FC-graphs inhibits GNNs from learning the most informative order of interactions. Such a phenomenon is broadly shared by several GNNs for different graph learning tasks and prevents GNNs from reaching the global minimum loss, so we name it a representation bottleneck. To overcome that, we propose a novel graph rewiring approach based on the pairwise interaction strengths to adjust the reception fields of each node dynamically. Extensive experiments in molecular property prediction and dynamic system forecast prove the superiority of our method over state-of-the-art GNN baselines. Besides, this paper provides a reasonable explanation of why subgraphs play a vital role in determining graph properties. The code is available at https://github.com/smiles724/bottleneck.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge