DeepNovoV2: Better de novo peptide sequencing with deep learning
Paper and Code
May 22, 2019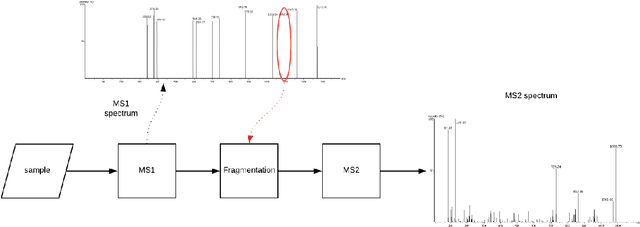
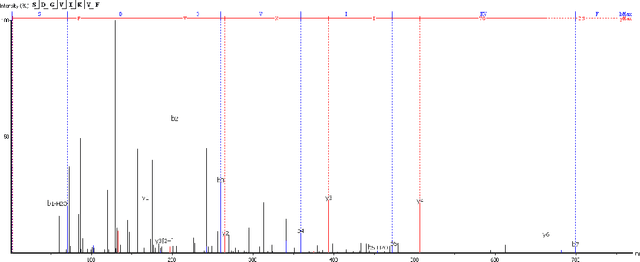
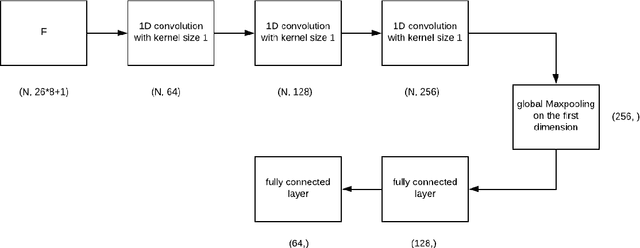
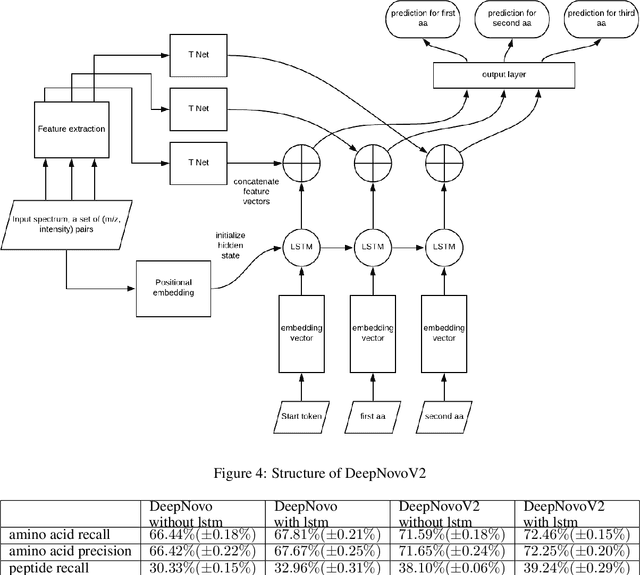
Personalized cancer vaccines are envisioned as the next generation rational cancer immunotherapy. The key step in developing personalized therapeutic cancer vaccines is to identify tumor-specific neoantigens that are on the surface of tumor cells. A promising method for this is through de novo peptide sequencing from mass spectrometry data. In this paper we introduce DeepNovoV2, the state-of-the-art model for peptide sequencing. In DeepNovoV2, a spectrum is directly represented as a set of (m/z, intensity) pairs, therefore it does not suffer from the accuracy-speed/memory trade-off problem. The model combines an order invariant network structure (T-Net) and recurrent neural networks and provides a complete end-to-end training and prediction framework to sequence patterns of peptides. Our experiments on a wide variety of data from different species show that DeepNovoV2 outperforms previous state-of-the-art methods, achieving 13.01-23.95\% higher accuracy at the peptide level.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge