CoNIC: Colon Nuclei Identification and Counting Challenge 2022
Paper and Code
Nov 29, 2021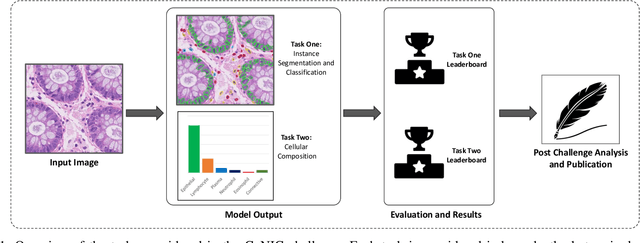
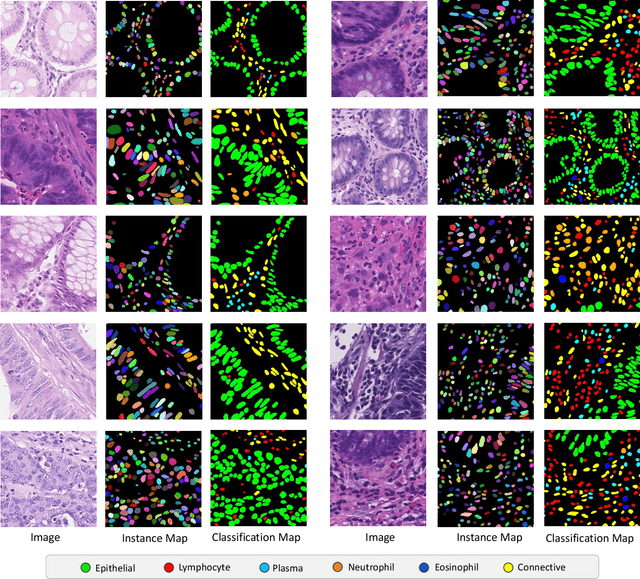
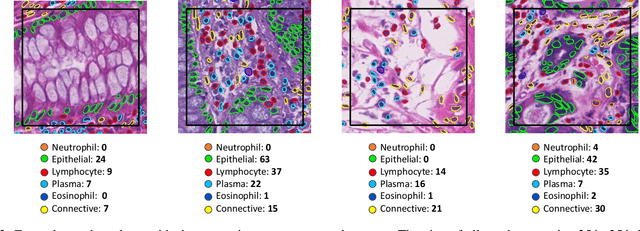
Nuclear segmentation, classification and quantification within Haematoxylin & Eosin stained histology images enables the extraction of interpretable cell-based features that can be used in downstream explainable models in computational pathology (CPath). However, automatic recognition of different nuclei is faced with a major challenge in that there are several different types of nuclei, some of them exhibiting large intra-class variability. To help drive forward research and innovation for automatic nuclei recognition in CPath, we organise the Colon Nuclei Identification and Counting (CoNIC) Challenge. The challenge encourages researchers to develop algorithms that perform segmentation, classification and counting of nuclei within the current largest known publicly available nuclei-level dataset in CPath, containing around half a million labelled nuclei. Therefore, the CoNIC challenge utilises over 10 times the number of nuclei as the previous largest challenge dataset for nuclei recognition. It is important for algorithms to be robust to input variation if we wish to deploy them in a clinical setting. Therefore, as part of this challenge we will also test the sensitivity of each submitted algorithm to certain input variations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge