AVP: Physics-informed Data Generation for Small-data Learning
Paper and Code
Feb 05, 2019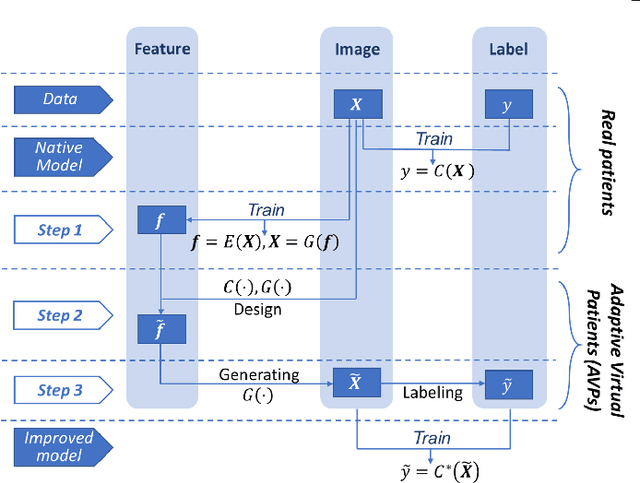
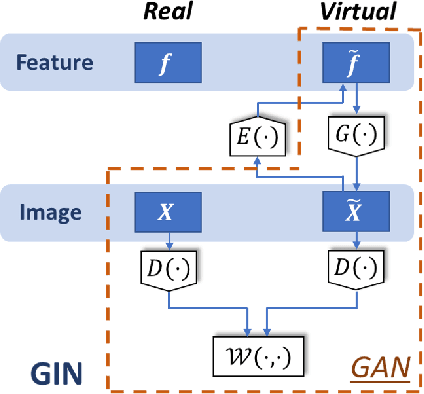

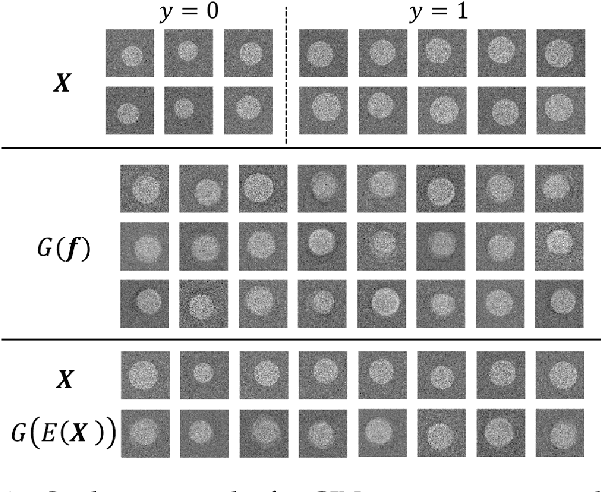
Deep neural networks have achieved great success in multiple learning problems, and attracted increasing attention from the medicine community. In reality, however, the limited availability and high costs of medical data is a major challenge of applying deep neural networks to computer-aided diagnosis and treatment planning. We address this challenge with adaptive virtual patients (AVPs) and the associated physics-informed learning framework. Specifically, the original training dataset is fused with an additional dataset of AVPs, which are generated by a data-driven model and the associated supervision (e.g., labels) is obtained by a physics-based approach. A key novelty in the proposed framework is the bidirectional and uncoupled generative invertible networks (GIN), which can extract pathophysiological features from the training medical image and generate pathophysiologically meaningful virtual patients. In order to mitigate the possibly high labeling cost of physical experiments, a $\mu$-measure design is conducted: this allows the AVPs to not only further explore the uncertain regions, but also balance the label distribution. We then discuss the pathophysiological interpretability of GIN both theoretically and experimentally, and demonstrate the effectiveness of AVPs using a real medical image dataset, in which the proposed AVPs lower the labeling cost by 90% while achieving a 15% improvement in prediction accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge