Auto-Prompting SAM for Mobile Friendly 3D Medical Image Segmentation
Paper and Code
Aug 28, 2023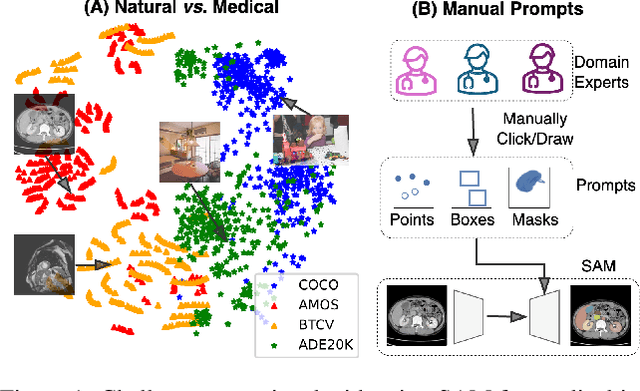
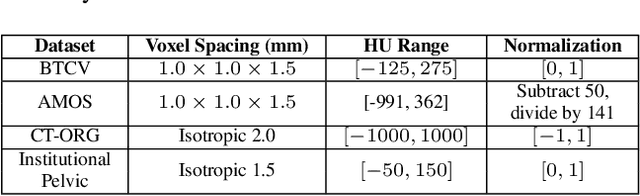
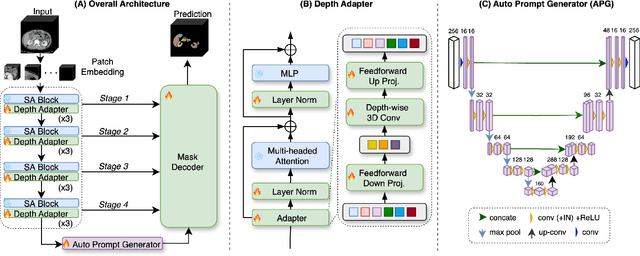
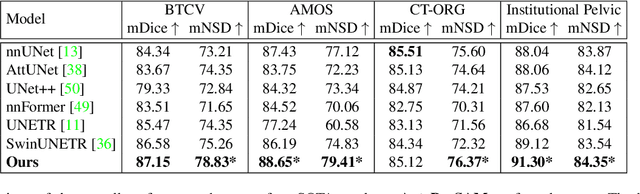
The Segment Anything Model (SAM) has rapidly been adopted for segmenting a wide range of natural images. However, recent studies have indicated that SAM exhibits subpar performance on 3D medical image segmentation tasks. In addition to the domain gaps between natural and medical images, disparities in the spatial arrangement between 2D and 3D images, the substantial computational burden imposed by powerful GPU servers, and the time-consuming manual prompt generation impede the extension of SAM to a broader spectrum of medical image segmentation applications. To address these challenges, in this work, we introduce a novel method, AutoSAM Adapter, designed specifically for 3D multi-organ CT-based segmentation. We employ parameter-efficient adaptation techniques in developing an automatic prompt learning paradigm to facilitate the transformation of the SAM model's capabilities to 3D medical image segmentation, eliminating the need for manually generated prompts. Furthermore, we effectively transfer the acquired knowledge of the AutoSAM Adapter to other lightweight models specifically tailored for 3D medical image analysis, achieving state-of-the-art (SOTA) performance on medical image segmentation tasks. Through extensive experimental evaluation, we demonstrate the AutoSAM Adapter as a critical foundation for effectively leveraging the emerging ability of foundation models in 2D natural image segmentation for 3D medical image segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge