Zilan Yu
CLLMFS: A Contrastive Learning enhanced Large Language Model Framework for Few-Shot Named Entity Recognition
Aug 23, 2024Abstract:Few-shot Named Entity Recognition (NER), the task of identifying named entities with only a limited amount of labeled data, has gained increasing significance in natural language processing. While existing methodologies have shown some effectiveness, such as enriching label semantics through various prompting modes or employing metric learning techniques, their performance exhibits limited robustness across diverse domains due to the lack of rich knowledge in their pre-trained models. To address this issue, we propose CLLMFS, a Contrastive Learning enhanced Large Language Model (LLM) Framework for Few-Shot Named Entity Recognition, achieving promising results with limited training data. Considering the impact of LLM's internal representations on downstream tasks, CLLMFS integrates Low-Rank Adaptation (LoRA) and contrastive learning mechanisms specifically tailored for few-shot NER. By enhancing the model's internal representations, CLLMFS effectively improves both entity boundary awareness ability and entity recognition accuracy. Our method has achieved state-of-the-art performance improvements on F1-score ranging from 2.58\% to 97.74\% over existing best-performing methods across several recognized benchmarks. Furthermore, through cross-domain NER experiments conducted on multiple datasets, we have further validated the robust generalization capability of our method. Our code will be released in the near future.
PtLnc-BXE: Prediction of plant lncRNAs using a Bagging-XGBoost-ensemble method with multiple features
Nov 01, 2019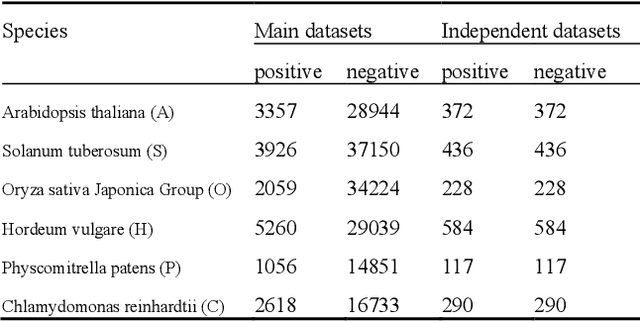
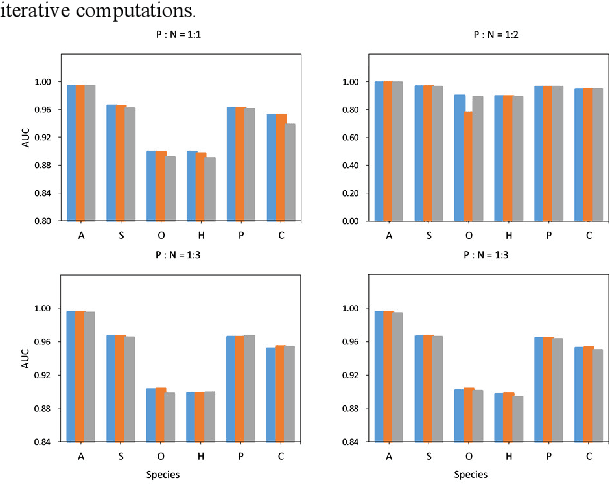
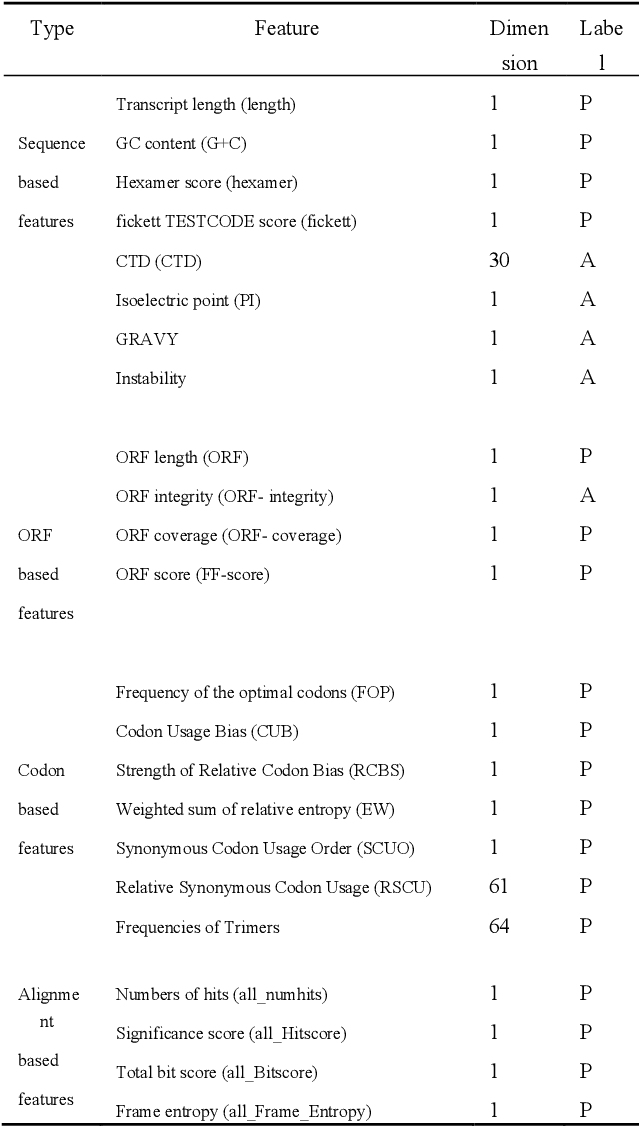
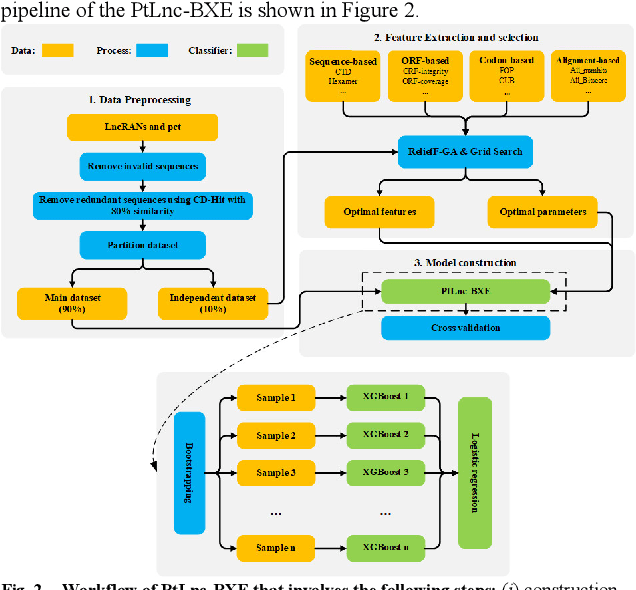
Abstract:Motivation: Long non-coding RNAs (lncRNAs) are a diverse class of RNA molecules with a length above 200 nucleotides that do not encode proteins. Since lncRNAs have involved in a wide range of functions in cellular and developmental processes, an increasing number of methods or tools for distin-guishing lncRNAs from coding RNAs have been proposed. However, most of the existing methods are designed for lncRNAs in animal systems, and only a few methods focus on the plant lncRNA identifica-tion. Different from lncRNAs in animal systems, plant lncRNAs have distinct characteristics. It is desira-ble to develop a computational method for accurate and rapid identification of plant lncRNAs. Results: Herein, we present a plant lncRNA prediction approach PtLnc-BXE, which combines multiple sequence features in two steps to develop an ensemble mode. First, a diverse number of plants lncRNA features are collected and filtered by feature selection and subsequently used to represent RNA se-quences. Then, the training dataset is sampled into several subsets using the bootstrapping technique, and base learners are constructed on data subsets by using XGBoost, and multiple base learners are further combined into a single meta-learner by using logistic regression. PtLnc-BXE outperformed other state-of-the-art plant lncRNA prediction methods, achieving higher AUC (> 95.9%) on the benchmark datasets. The studies across different plant species reveal that the different species have a high overlap between their selected features for modeling. Therefore, it is possible to build the cross-species predic-tion models for plant lncRNAs. Availability: The scripts and data can be downloaded at https://github.com/xxxxx Contact: example@example.org Supplementary information: Supplementary data are available at Bioinformatics online.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge