Zhuxian Guo
Lymphoid Infiltration Assessment of the Tumor Margins in H&E Slides
Jul 23, 2024



Abstract:Lymphoid infiltration at tumor margins is a key prognostic marker in solid tumors, playing a crucial role in guiding immunotherapy decisions. Current assessment methods, heavily reliant on immunohistochemistry (IHC), face challenges in tumor margin delineation and are affected by tissue preservation conditions. In contrast, we propose a Hematoxylin and Eosin (H&E) staining-based approach, underpinned by an advanced lymphocyte segmentation model trained on a public dataset for the precise detection of CD3+ and CD20+ lymphocytes. In our colorectal cancer study, we demonstrate that our H&E-based method offers a compelling alternative to traditional IHC, achieving comparable results in many cases. Our method's validity is further explored through a Turing test, involving blinded assessments by a pathologist of anonymized curves from H&E and IHC slides. This approach invites the medical community to consider Turing tests as a standard for evaluating medical applications involving expert human evaluation, thereby opening new avenues for enhancing cancer management and immunotherapy planning.
Optimizing Lymphocyte Detection in Breast Cancer Whole Slide Imaging through Data-Centric Strategies
May 22, 2024
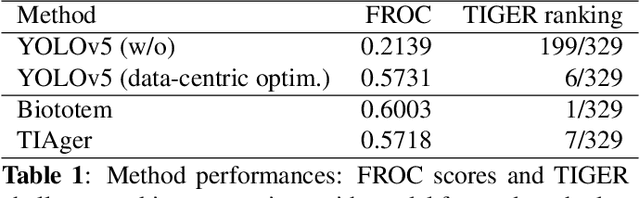


Abstract:Efficient and precise quantification of lymphocytes in histopathology slides is imperative for the characterization of the tumor microenvironment and immunotherapy response insights. We developed a data-centric optimization pipeline that attain great lymphocyte detection performance using an off-the-shelf YOLOv5 model, without any architectural modifications. Our contribution that rely on strategic dataset augmentation strategies, includes novel biological upsampling and custom visual cohesion transformations tailored to the unique properties of tissue imagery, and enables to dramatically improve model performances. Our optimization reveals a pivotal realization: given intensive customization, standard computational pathology models can achieve high-capability biomarker development, without increasing the architectural complexity. We showcase the interest of this approach in the context of breast cancer where our strategies lead to good lymphocyte detection performances, echoing a broadly impactful paradigm shift. Furthermore, our data curation techniques enable crucial histological analysis benchmarks, highlighting improved generalizable potential.
A Hierarchical Transformer Encoder to Improve Entire Neoplasm Segmentation on Whole Slide Image of Hepatocellular Carcinoma
Jul 11, 2023



Abstract:In digital histopathology, entire neoplasm segmentation on Whole Slide Image (WSI) of Hepatocellular Carcinoma (HCC) plays an important role, especially as a preprocessing filter to automatically exclude healthy tissue, in histological molecular correlations mining and other downstream histopathological tasks. The segmentation task remains challenging due to HCC's inherent high-heterogeneity and the lack of dependency learning in large field of view. In this article, we propose a novel deep learning architecture with a hierarchical Transformer encoder, HiTrans, to learn the global dependencies within expanded 4096$\times$4096 WSI patches. HiTrans is designed to encode and decode the patches with larger reception fields and the learned global dependencies, compared to the state-of-the-art Fully Convolutional Neural networks (FCNN). Empirical evaluations verified that HiTrans leads to better segmentation performance by taking into account regional and global dependency information.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge