Zhenhuan Zhou
Deep Learning in Dental Image Analysis: A Systematic Review of Datasets, Methodologies, and Emerging Challenges
Oct 23, 2025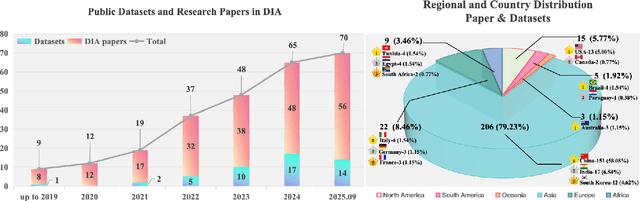
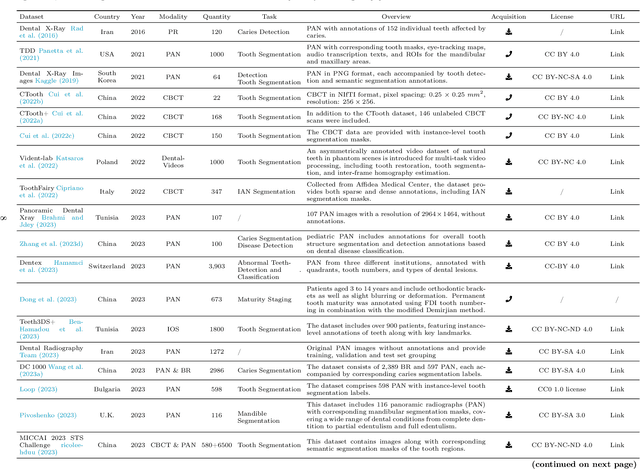
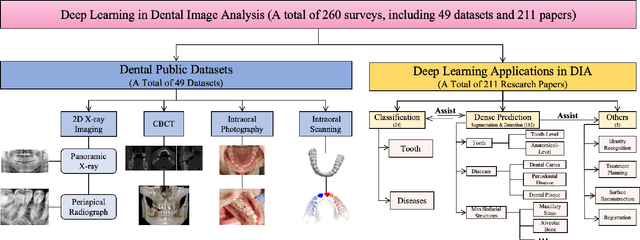
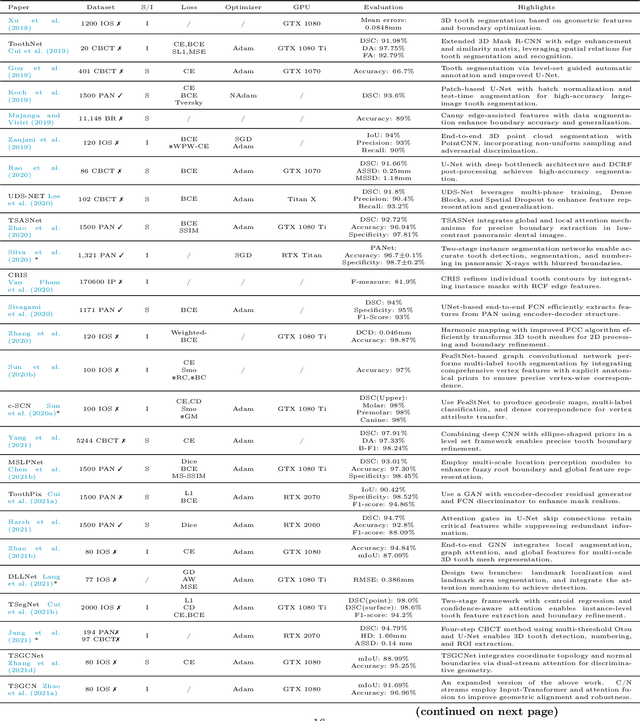
Abstract:Efficient analysis and processing of dental images are crucial for dentists to achieve accurate diagnosis and optimal treatment planning. However, dental imaging inherently poses several challenges, such as low contrast, metallic artifacts, and variations in projection angles. Combined with the subjectivity arising from differences in clinicians' expertise, manual interpretation often proves time-consuming and prone to inconsistency. Artificial intelligence (AI)-based automated dental image analysis (DIA) offers a promising solution to these issues and has become an integral part of computer-aided dental diagnosis and treatment. Among various AI technologies, deep learning (DL) stands out as the most widely applied and influential approach due to its superior feature extraction and representation capabilities. To comprehensively summarize recent progress in this field, we focus on the two fundamental aspects of DL research-datasets and models. In this paper, we systematically review 260 studies on DL applications in DIA, including 49 papers on publicly available dental datasets and 211 papers on DL-based algorithms. We first introduce the basic concepts of dental imaging and summarize the characteristics and acquisition methods of existing datasets. Then, we present the foundational techniques of DL and categorize relevant models and algorithms according to different DIA tasks, analyzing their network architectures, optimization strategies, training methods, and performance. Furthermore, we summarize commonly used training and evaluation metrics in the DIA domain. Finally, we discuss the current challenges of existing research and outline potential future directions. We hope that this work provides a valuable and systematic reference for researchers in this field. All supplementary materials and detailed comparison tables will be made publicly available on GitHub.
Med-URWKV: Pure RWKV With ImageNet Pre-training For Medical Image Segmentation
Jun 12, 2025Abstract:Medical image segmentation is a fundamental and key technology in computer-aided diagnosis and treatment. Previous methods can be broadly classified into three categories: convolutional neural network (CNN) based, Transformer based, and hybrid architectures that combine both. However, each of them has its own limitations, such as restricted receptive fields in CNNs or the computational overhead caused by the quadratic complexity of Transformers. Recently, the Receptance Weighted Key Value (RWKV) model has emerged as a promising alternative for various vision tasks, offering strong long-range modeling capabilities with linear computational complexity. Some studies have also adapted RWKV to medical image segmentation tasks, achieving competitive performance. However, most of these studies focus on modifications to the Vision-RWKV (VRWKV) mechanism and train models from scratch, without exploring the potential advantages of leveraging pre-trained VRWKV models for medical image segmentation tasks. In this paper, we propose Med-URWKV, a pure RWKV-based architecture built upon the U-Net framework, which incorporates ImageNet-based pretraining to further explore the potential of RWKV in medical image segmentation tasks. To the best of our knowledge, Med-URWKV is the first pure RWKV segmentation model in the medical field that can directly reuse a large-scale pre-trained VRWKV encoder. Experimental results on seven datasets demonstrate that Med-URWKV achieves comparable or even superior segmentation performance compared to other carefully optimized RWKV models trained from scratch. This validates the effectiveness of using a pretrained VRWKV encoder in enhancing model performance. The codes will be released.
Cross-Frequency Collaborative Training Network and Dataset for Semi-supervised First Molar Root Canal Segmentation
Apr 16, 2025Abstract:Root canal (RC) treatment is a highly delicate and technically complex procedure in clinical practice, heavily influenced by the clinicians' experience and subjective judgment. Deep learning has made significant advancements in the field of computer-aided diagnosis (CAD) because it can provide more objective and accurate diagnostic results. However, its application in RC treatment is still relatively rare, mainly due to the lack of public datasets in this field. To address this issue, in this paper, we established a First Molar Root Canal segmentation dataset called FMRC-2025. Additionally, to alleviate the workload of manual annotation for dentists and fully leverage the unlabeled data, we designed a Cross-Frequency Collaborative training semi-supervised learning (SSL) Network called CFC-Net. It consists of two components: (1) Cross-Frequency Collaborative Mean Teacher (CFC-MT), which introduces two specialized students (SS) and one comprehensive teacher (CT) for collaborative multi-frequency training. The CT and SS are trained on different frequency components while fully integrating multi-frequency knowledge through cross and full frequency consistency supervisions. (2) Uncertainty-guided Cross-Frequency Mix (UCF-Mix) mechanism enables the network to generate high-confidence pseudo-labels while learning to integrate multi-frequency information and maintaining the structural integrity of the targets. Extensive experiments on FMRC-2025 and three public dental datasets demonstrate that CFC-MT is effective for RC segmentation and can also exhibit strong generalizability on other dental segmentation tasks, outperforming state-of-the-art SSL medical image segmentation methods. Codes and dataset will be released.
PRAD: Periapical Radiograph Analysis Dataset and Benchmark Model Development
Apr 10, 2025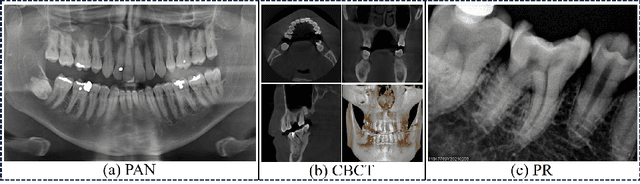
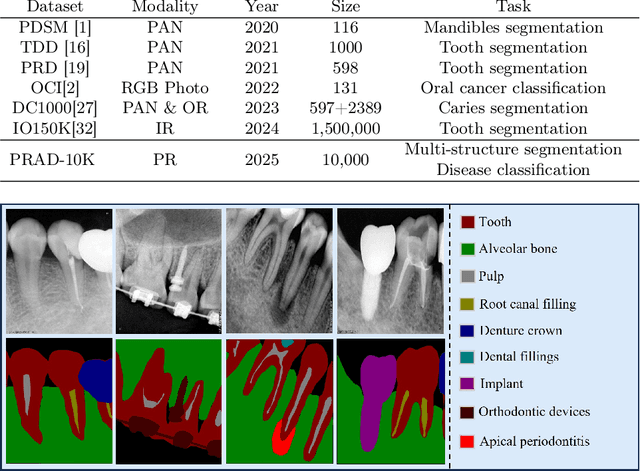
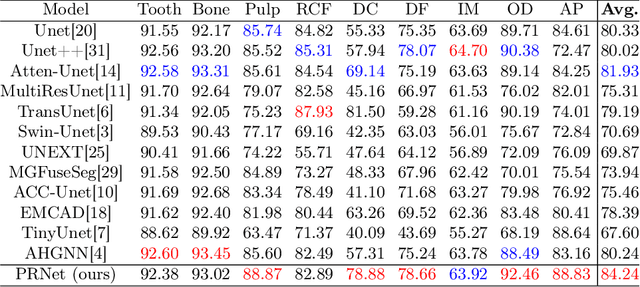
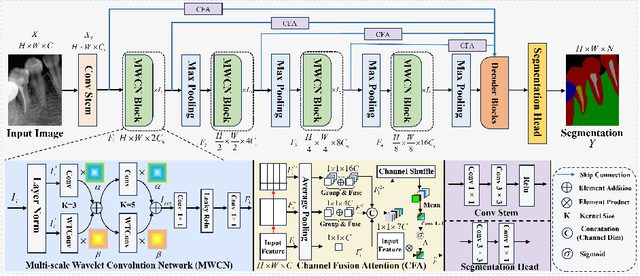
Abstract:Deep learning (DL), a pivotal technology in artificial intelligence, has recently gained substantial traction in the domain of dental auxiliary diagnosis. However, its application has predominantly been confined to imaging modalities such as panoramic radiographs and Cone Beam Computed Tomography, with limited focus on auxiliary analysis specifically targeting Periapical Radiographs (PR). PR are the most extensively utilized imaging modality in endodontics and periodontics due to their capability to capture detailed local lesions at a low cost. Nevertheless, challenges such as resolution limitations and artifacts complicate the annotation and recognition of PR, leading to a scarcity of publicly available, large-scale, high-quality PR analysis datasets. This scarcity has somewhat impeded the advancement of DL applications in PR analysis. In this paper, we present PRAD-10K, a dataset for PR analysis. PRAD-10K comprises 10,000 clinical periapical radiograph images, with pixel-level annotations provided by professional dentists for nine distinct anatomical structures, lesions, and artificial restorations or medical devices, We also include classification labels for images with typical conditions or lesions. Furthermore, we introduce a DL network named PRNet to establish benchmarks for PR segmentation tasks. Experimental results demonstrate that PRNet surpasses previous state-of-the-art medical image segmentation models on the PRAD-10K dataset. The codes and dataset will be made publicly available.
Spatial-Frequency Dual Progressive Attention Network For Medical Image Segmentation
Jun 12, 2024Abstract:In medical images, various types of lesions often manifest significant differences in their shape and texture. Accurate medical image segmentation demands deep learning models with robust capabilities in multi-scale and boundary feature learning. However, previous networks still have limitations in addressing the above issues. Firstly, previous networks simultaneously fuse multi-level features or employ deep supervision to enhance multi-scale learning. However, this may lead to feature redundancy and excessive computational overhead, which is not conducive to network training and clinical deployment. Secondly, the majority of medical image segmentation networks exclusively learn features in the spatial domain, disregarding the abundant global information in the frequency domain. This results in a bias towards low-frequency components, neglecting crucial high-frequency information. To address these problems, we introduce SF-UNet, a spatial-frequency dual-domain attention network. It comprises two main components: the Multi-scale Progressive Channel Attention (MPCA) block, which progressively extract multi-scale features across adjacent encoder layers, and the lightweight Frequency-Spatial Attention (FSA) block, with only 0.05M parameters, enabling concurrent learning of texture and boundary features from both spatial and frequency domains. We validate the effectiveness of the proposed SF-UNet on three public datasets. Experimental results show that compared to previous state-of-the-art (SOTA) medical image segmentation networks, SF-UNet achieves the best performance, and achieves up to 9.4\% and 10.78\% improvement in DSC and IOU. Codes will be released at https://github.com/nkicsl/SF-UNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge