Zhaohui Qin
Deep Causal Generative Models with Property Control
May 25, 2024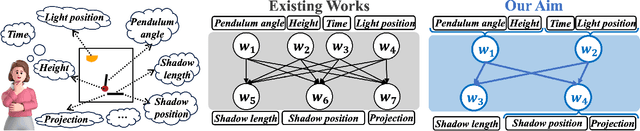
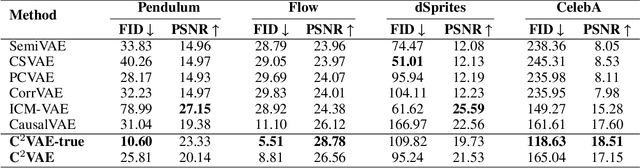
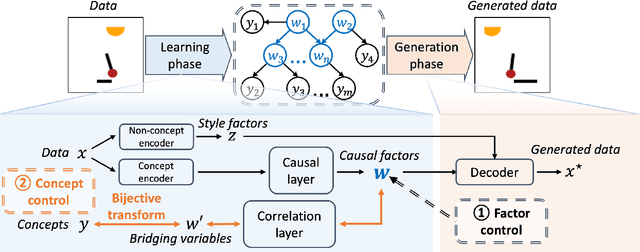
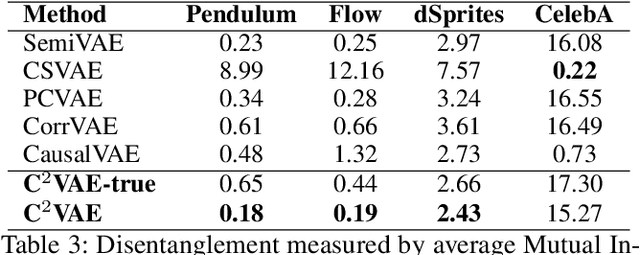
Abstract:Generating data with properties of interest by external users while following the right causation among its intrinsic factors is important yet has not been well addressed jointly. This is due to the long-lasting challenge of jointly identifying key latent variables, their causal relations, and their correlation with properties of interest, as well as how to leverage their discoveries toward causally controlled data generation. To address these challenges, we propose a novel deep generative framework called the Correlation-aware Causal Variational Auto-encoder (C2VAE). This framework simultaneously recovers the correlation and causal relationships between properties using disentangled latent vectors. Specifically, causality is captured by learning the causal graph on latent variables through a structural causal model, while correlation is learned via a novel correlation pooling algorithm. Extensive experiments demonstrate C2VAE's ability to accurately recover true causality and correlation, as well as its superiority in controllable data generation compared to baseline models.
Gene-associated Disease Discovery Powered by Large Language Models
Jan 16, 2024Abstract:The intricate relationship between genetic variation and human diseases has been a focal point of medical research, evidenced by the identification of risk genes regarding specific diseases. The advent of advanced genome sequencing techniques has significantly improved the efficiency and cost-effectiveness of detecting these genetic markers, playing a crucial role in disease diagnosis and forming the basis for clinical decision-making and early risk assessment. To overcome the limitations of existing databases that record disease-gene associations from existing literature, which often lack real-time updates, we propose a novel framework employing Large Language Models (LLMs) for the discovery of diseases associated with specific genes. This framework aims to automate the labor-intensive process of sifting through medical literature for evidence linking genetic variations to diseases, thereby enhancing the efficiency of disease identification. Our approach involves using LLMs to conduct literature searches, summarize relevant findings, and pinpoint diseases related to specific genes. This paper details the development and application of our LLM-powered framework, demonstrating its potential in streamlining the complex process of literature retrieval and summarization to identify diseases associated with specific genetic variations.
Domain Generalization Deep Graph Transformation
May 23, 2023Abstract:Graph transformation that predicts graph transition from one mode to another is an important and common problem. Despite much progress in developing advanced graph transformation techniques in recent years, the fundamental assumption typically required in machine-learning models that the testing and training data preserve the same distribution does not always hold. As a result, domain generalization graph transformation that predicts graphs not available in the training data is under-explored, with multiple key challenges to be addressed including (1) the extreme space complexity when training on all input-output mode combinations, (2) difference of graph topologies between the input and the output modes, and (3) how to generalize the model to (unseen) target domains that are not in the training data. To fill the gap, we propose a multi-input, multi-output, hypernetwork-based graph neural network (MultiHyperGNN) that employs a encoder and a decoder to encode topologies of both input and output modes and semi-supervised link prediction to enhance the graph transformation task. Instead of training on all mode combinations, MultiHyperGNN preserves a constant space complexity with the encoder and the decoder produced by two novel hypernetworks. Comprehensive experiments show that MultiHyperGNN has a superior performance than competing models in both prediction and domain generalization tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge