Yusong Li
Optimizing Medical Image Segmentation with Advanced Decoder Design
Oct 05, 2024



Abstract:U-Net is widely used in medical image segmentation due to its simple and flexible architecture design. To address the challenges of scale and complexity in medical tasks, several variants of U-Net have been proposed. In particular, methods based on Vision Transformer (ViT), represented by Swin UNETR, have gained widespread attention in recent years. However, these improvements often focus on the encoder, overlooking the crucial role of the decoder in optimizing segmentation details. This design imbalance limits the potential for further enhancing segmentation performance. To address this issue, we analyze the roles of various decoder components, including upsampling method, skip connection, and feature extraction module, as well as the shortcomings of existing methods. Consequently, we propose Swin DER (i.e., Swin UNETR Decoder Enhanced and Refined) by specifically optimizing the design of these three components. Swin DER performs upsampling using learnable interpolation algorithm called offset coordinate neighborhood weighted up sampling (Onsampling) and replaces traditional skip connection with spatial-channel parallel attention gate (SCP AG). Additionally, Swin DER introduces deformable convolution along with attention mechanism in the feature extraction module of the decoder. Our model design achieves excellent results, surpassing other state-of-the-art methods on both the Synapse and the MSD brain tumor segmentation task. Code is available at: https://github.com/WillBeanYang/Swin-DER
More complex encoder is not all you need
Sep 21, 2023



Abstract:U-Net and its variants have been widely used in medical image segmentation. However, most current U-Net variants confine their improvement strategies to building more complex encoder, while leaving the decoder unchanged or adopting a simple symmetric structure. These approaches overlook the true functionality of the decoder: receiving low-resolution feature maps from the encoder and restoring feature map resolution and lost information through upsampling. As a result, the decoder, especially its upsampling component, plays a crucial role in enhancing segmentation outcomes. However, in 3D medical image segmentation, the commonly used transposed convolution can result in visual artifacts. This issue stems from the absence of direct relationship between adjacent pixels in the output feature map. Furthermore, plain encoder has already possessed sufficient feature extraction capability because downsampling operation leads to the gradual expansion of the receptive field, but the loss of information during downsampling process is unignorable. To address the gap in relevant research, we extend our focus beyond the encoder and introduce neU-Net (i.e., not complex encoder U-Net), which incorporates a novel Sub-pixel Convolution for upsampling to construct a powerful decoder. Additionally, we introduce multi-scale wavelet inputs module on the encoder side to provide additional information. Our model design achieves excellent results, surpassing other state-of-the-art methods on both the Synapse and ACDC datasets.
Inferring Gene Regulatory Neural Networks for Bacterial Decision Making in Biofilms
Jan 10, 2023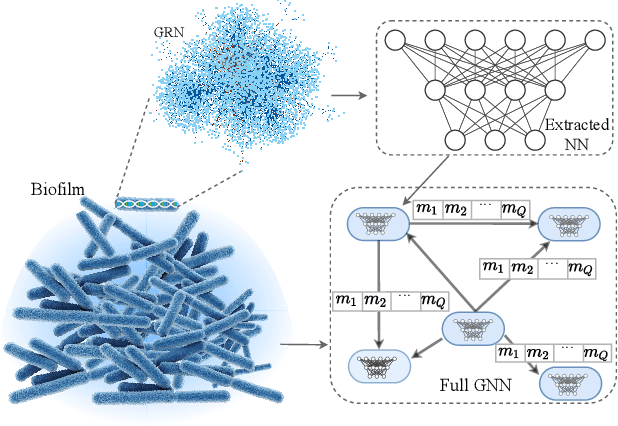
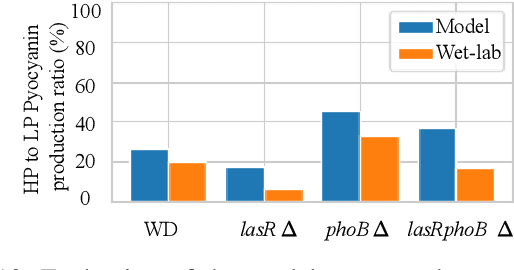
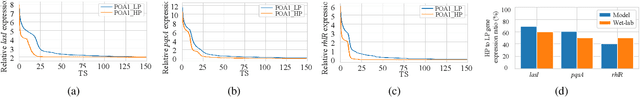
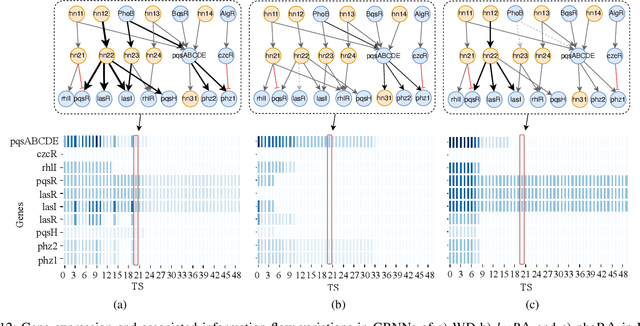
Abstract:Bacterial cells are sensitive to a range of external signals used to learn the environment. These incoming external signals are then processed using a Gene Regulatory Network (GRN), exhibiting similarities to modern computing algorithms. An in-depth analysis of gene expression dynamics suggests an inherited Gene Regulatory Neural Network (GRNN) behavior within the GRN that enables the cellular decision-making based on received signals from the environment and neighbor cells. In this study, we extract a sub-network of \textit{Pseudomonas aeruginosa} GRN that is associated with one virulence factor: pyocyanin production as a use case to investigate the GRNN behaviors. Further, using Graph Neural Network (GNN) architecture, we model a single species biofilm to reveal the role of GRNN dynamics on ecosystem-wide decision-making. Varying environmental conditions, we prove that the extracted GRNN computes input signals similar to natural decision-making process of the cell. Identifying of neural network behaviors in GRNs may lead to more accurate bacterial cell activity predictive models for many applications, including human health-related problems and agricultural applications. Further, this model can produce data on causal relationships throughout the network, enabling the possibility of designing tailor-made infection-controlling mechanisms. More interestingly, these GRNNs can perform computational tasks for bio-hybrid computing systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge