Yuhang Sheng
Two-Layer Reinforcement Learning-Assisted Joint Beamforming and Trajectory Optimization for Multi-UAV Downlink Communications
Jan 19, 2026Abstract:Unmanned aerial vehicles (UAVs) are pivotal for future 6G non-terrestrial networks, yet their high mobility creates a complex coupled optimization problem for beamforming and trajectory design. Existing numerical methods suffer from prohibitive latency, while standard deep learning often ignores dynamic interference topology, limiting scalability. To address these issues, this paper proposes a hierarchically decoupled framework synergizing graph neural networks (GNNs) with multi-agent reinforcement learning. Specifically, on the short timescale, we develop a topology-aware GNN beamformer by incorporating GraphNorm. By modeling the dynamic UAV-user association as a time-varying heterogeneous graph, this method explicitly extracts interference patterns to achieve sub-millisecond inference. On the long timescale, trajectory planning is modeled as a decentralized partially observable Markov decision process and solved via the multi-agent proximal policy optimization algorithm under the centralized training with decentralized execution paradigm, facilitating cooperative behaviors. Extensive simulation results demonstrate that the proposed framework significantly outperforms state-of-the-art optimization heuristics and deep learning baselines in terms of system sum rate, convergence speed, and generalization capability.
Subtype-Former: a deep learning approach for cancer subtype discovery with multi-omics data
Jul 28, 2022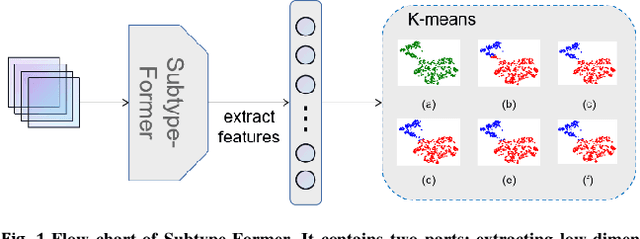
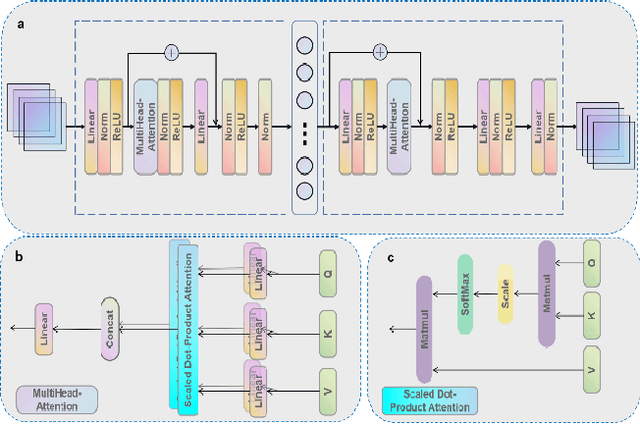
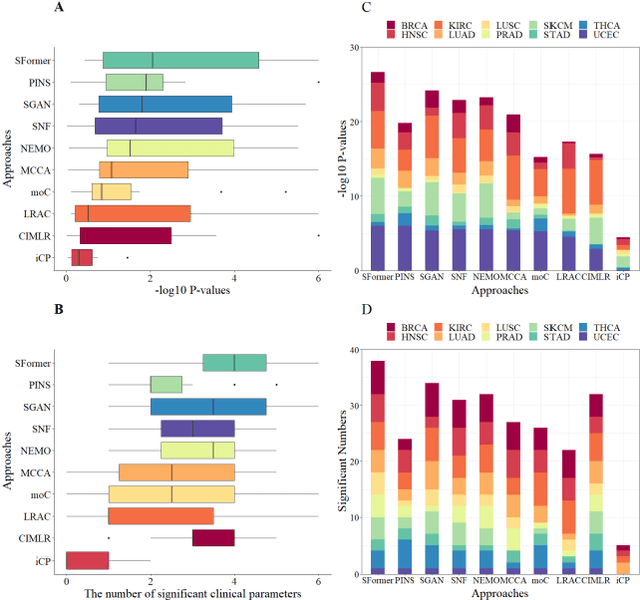
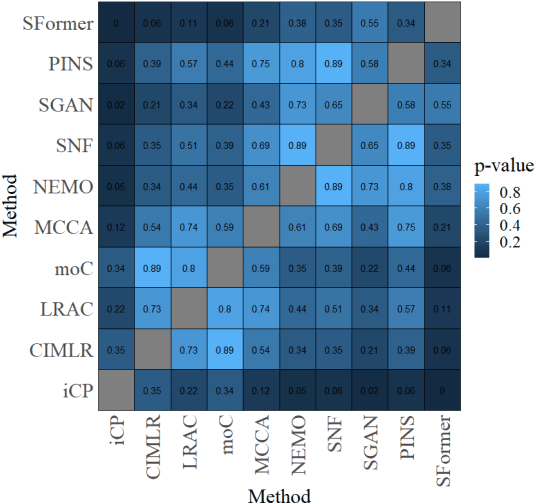
Abstract:Motivation: Cancer is heterogeneous, affecting the precise approach to personalized treatment. Accurate subtyping can lead to better survival rates for cancer patients. High-throughput technologies provide multiple omics data for cancer subtyping. However, precise cancer subtyping remains challenging due to the large amount and high dimensionality of omics data. Results: This study proposed Subtype-Former, a deep learning method based on MLP and Transformer Block, to extract the low-dimensional representation of the multi-omics data. K-means and Consensus Clustering are also used to achieve accurate subtyping results. We compared Subtype-Former with the other state-of-the-art subtyping methods across the TCGA 10 cancer types. We found that Subtype-Former can perform better on the benchmark datasets of more than 5000 tumors based on the survival analysis. In addition, Subtype-Former also achieved outstanding results in pan-cancer subtyping, which can help analyze the commonalities and differences across various cancer types at the molecular level. Finally, we applied Subtype-Former to the TCGA 10 types of cancers. We identified 50 essential biomarkers, which can be used to study targeted cancer drugs and promote the development of cancer treatments in the era of precision medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge