Yufeng Jiang
DeepMoLM: Leveraging Visual and Geometric Structural Information for Molecule-Text Modeling
Jan 21, 2026Abstract:AI models for drug discovery and chemical literature mining must interpret molecular images and generate outputs consistent with 3D geometry and stereochemistry. Most molecular language models rely on strings or graphs, while vision-language models often miss stereochemical details and struggle to map continuous 3D structures into discrete tokens. We propose DeepMoLM: Deep Molecular Language M odeling, a dual-view framework that grounds high-resolution molecular images in geometric invariants derived from molecular conformations. DeepMoLM preserves high-frequency evidence from 1024 $\times$ 1024 inputs, encodes conformer neighborhoods as discrete Extended 3-Dimensional Fingerprints, and fuses visual and geometric streams with cross-attention, enabling physically grounded generation without atom coordinates. DeepMoLM improves PubChem captioning with a 12.3% relative METEOR gain over the strongest generalist baseline while staying competitive with specialist methods. It produces valid numeric outputs for all property queries and attains MAE 13.64 g/mol on Molecular Weight and 37.89 on Complexity in the specialist setting. On ChEBI-20 description generation from images, it exceeds generalist baselines and matches state-of- the-art vision-language models. Code is available at https://github.com/1anj/DeepMoLM.
Structure-Aware Contrastive Learning with Fine-Grained Binding Representations for Drug Discovery
Sep 18, 2025



Abstract:Accurate identification of drug-target interactions (DTI) remains a central challenge in computational pharmacology, where sequence-based methods offer scalability. This work introduces a sequence-based drug-target interaction framework that integrates structural priors into protein representations while maintaining high-throughput screening capability. Evaluated across multiple benchmarks, the model achieves state-of-the-art performance on Human and BioSNAP datasets and remains competitive on BindingDB. In virtual screening tasks, it surpasses prior methods on LIT-PCBA, yielding substantial gains in AUROC and BEDROC. Ablation studies confirm the critical role of learned aggregation, bilinear attention, and contrastive alignment in enhancing predictive robustness. Embedding visualizations reveal improved spatial correspondence with known binding pockets and highlight interpretable attention patterns over ligand-residue contacts. These results validate the framework's utility for scalable and structure-aware DTI prediction.
MLLA-UNet: Mamba-like Linear Attention in an Efficient U-Shape Model for Medical Image Segmentation
Oct 31, 2024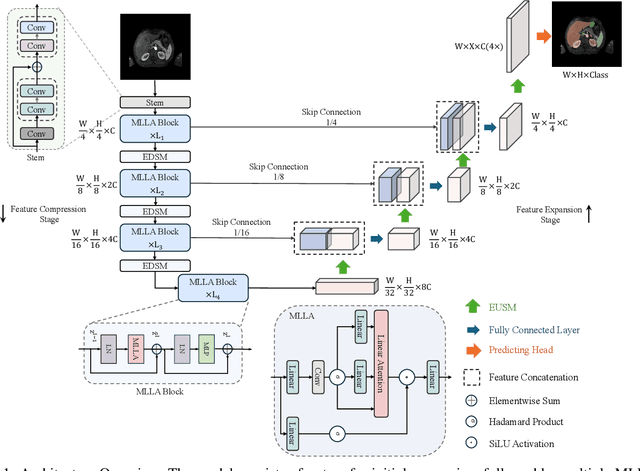
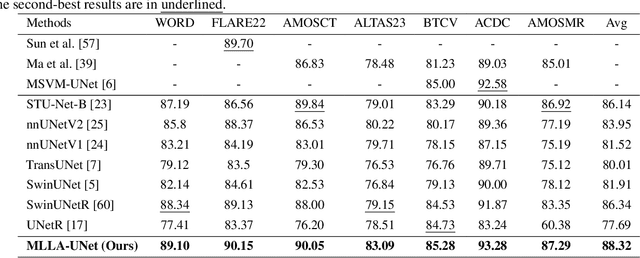
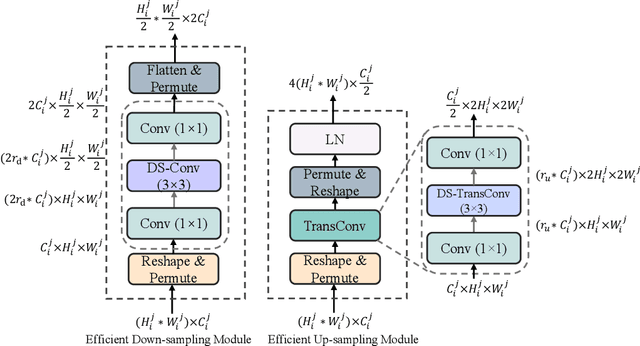
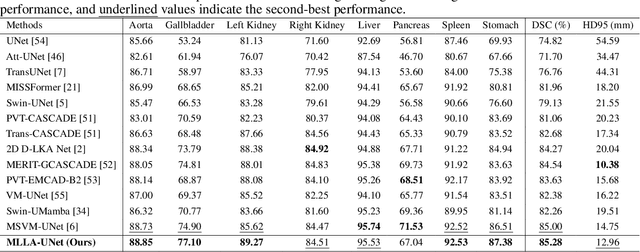
Abstract:Recent advancements in medical imaging have resulted in more complex and diverse images, with challenges such as high anatomical variability, blurred tissue boundaries, low organ contrast, and noise. Traditional segmentation methods struggle to address these challenges, making deep learning approaches, particularly U-shaped architectures, increasingly prominent. However, the quadratic complexity of standard self-attention makes Transformers computationally prohibitive for high-resolution images. To address these challenges, we propose MLLA-UNet (Mamba-Like Linear Attention UNet), a novel architecture that achieves linear computational complexity while maintaining high segmentation accuracy through its innovative combination of linear attention and Mamba-inspired adaptive mechanisms, complemented by an efficient symmetric sampling structure for enhanced feature processing. Our architecture effectively preserves essential spatial features while capturing long-range dependencies at reduced computational complexity. Additionally, we introduce a novel sampling strategy for multi-scale feature fusion. Experiments demonstrate that MLLA-UNet achieves state-of-the-art performance on six challenging datasets with 24 different segmentation tasks, including but not limited to FLARE22, AMOS CT, and ACDC, with an average DSC of 88.32%. These results underscore the superiority of MLLA-UNet over existing methods. Our contributions include the novel 2D segmentation architecture and its empirical validation. The code is available via https://github.com/csyfjiang/MLLA-UNet.
M$^4$oE: A Foundation Model for Medical Multimodal Image Segmentation with Mixture of Experts
May 15, 2024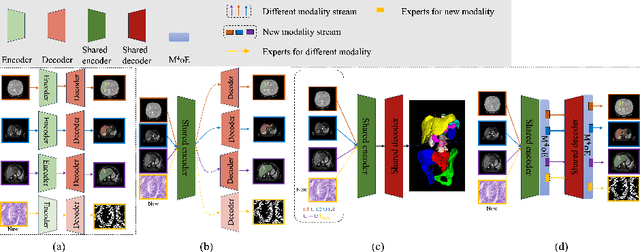
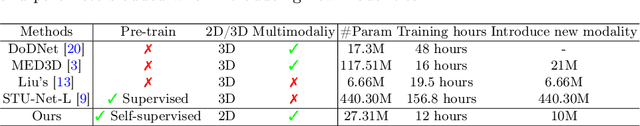
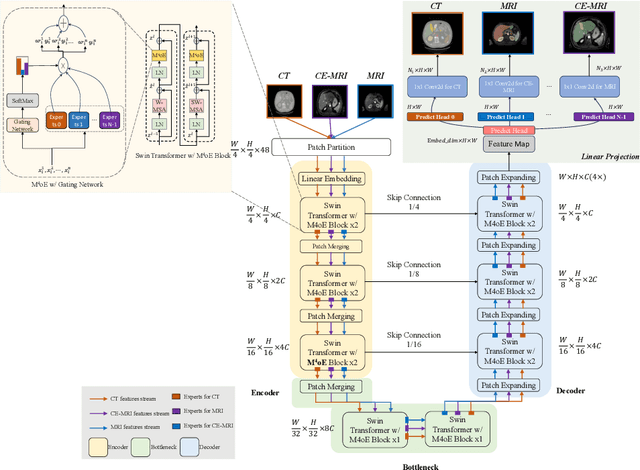
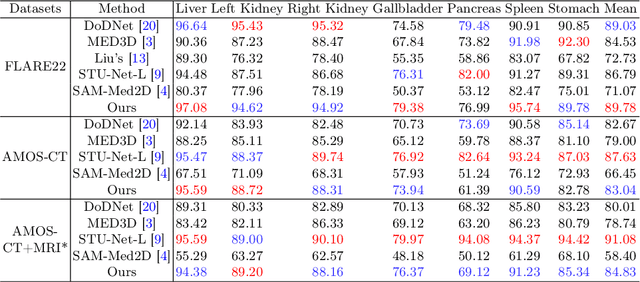
Abstract:Medical imaging data is inherently heterogeneous across different modalities and clinical centers, posing unique challenges for developing generalizable foundation models. Conventional entails training distinct models per dataset or using a shared encoder with modality-specific decoders. However, these approaches incur heavy computational overheads and suffer from poor scalability. To address these limitations, we propose the Medical Multimodal Mixture of Experts (M$^4$oE) framework, leveraging the SwinUNet architecture. Specifically, M$^4$oE comprises modality-specific experts; each separately initialized to learn features encoding domain knowledge. Subsequently, a gating network is integrated during fine-tuning to modulate each expert's contribution to the collective predictions dynamically. This enhances model interpretability and generalization ability while retaining expertise specialization. Simultaneously, the M$^4$oE architecture amplifies the model's parallel processing capabilities, and it also ensures the model's adaptation to new modalities with ease. Experiments across three modalities reveal that M$^4$oE can achieve 3.45% over STU-Net-L, 5.11% over MED3D, and 11.93% over SAM-Med2D across the MICCAI FLARE22, AMOS2022, and ATLAS2023 datasets. Moreover, M$^4$oE showcases a significant reduction in training duration with 7 hours less while maintaining a parameter count that is only 30% of its compared methods. The code is available at https://github.com/JefferyJiang-YF/M4oE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge