Yilin Leng
for the Alzheimer's Disease Neuroimaging Initiative
Adaptive Critical Subgraph Mining for Cognitive Impairment Conversion Prediction with T1-MRI-based Brain Network
Mar 20, 2024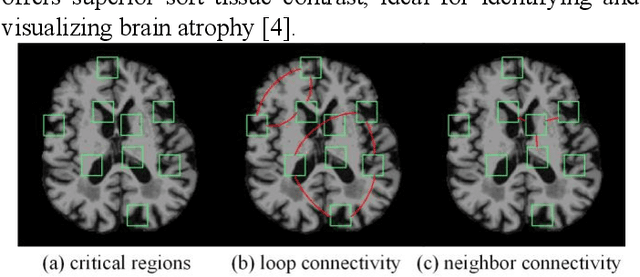
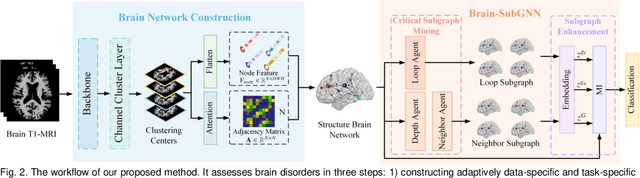
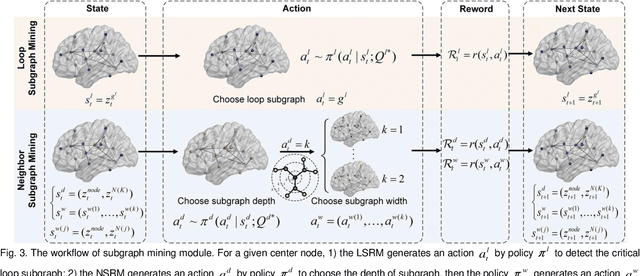
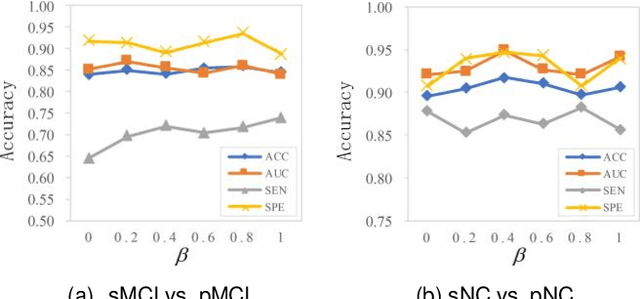
Abstract:Prediction the conversion to early-stage dementia is critical for mitigating its progression but remains challenging due to subtle cognitive impairments and structural brain changes. Traditional T1-weighted magnetic resonance imaging (T1-MRI) research focus on identifying brain atrophy regions but often fails to address the intricate connectivity between them. This limitation underscores the necessity of focuing on inter-regional connectivity for a comprehensive understand of the brain's complex network. Moreover, there is a pressing demand for methods that adaptively preserve and extract critical information, particularly specialized subgraph mining techniques for brain networks. These are essential for developing high-quality feature representations that reveal critical spatial impacts of structural brain changes and its topology. In this paper, we propose Brain-SubGNN, a novel graph representation network to mine and enhance critical subgraphs based on T1-MRI. This network provides a subgraph-level interpretation, enhancing interpretability and insights for graph analysis. The process begins by extracting node features and a correlation matrix between nodes to construct a task-oriented brain network. Brain-SubGNN then adaptively identifies and enhances critical subgraphs, capturing both loop and neighbor subgraphs. This method reflects the loop topology and local changes, indicative of long-range connections, and maintains local and global brain attributes. Extensive experiments validate the effectiveness and advantages of Brain-SubGNN, demonstrating its potential as a powerful tool for understanding and diagnosing early-stage dementia. Source code is available at https://github.com/Leng-10/Brain-SubGNN.
Dynamic Structural Brain Network Construction by Hierarchical Prototype Embedding GCN using T1-MRI
May 17, 2023Abstract:Constructing structural brain networks using T1-weighted magnetic resonance imaging (T1-MRI) presents a significant challenge due to the lack of direct regional connectivity information. Current methods with T1-MRI rely on predefined regions or isolated pretrained location modules to obtain atrophic regions, which neglects individual specificity. Besides, existing methods capture global structural context only on the whole-image-level, which weaken correlation between regions and the hierarchical distribution nature of brain connectivity.We hereby propose a novel dynamic structural brain network construction method based on T1-MRI, which can dynamically localize critical regions and constrain the hierarchical distribution among them for constructing dynamic structural brain network. Specifically, we first cluster spatially-correlated channel and generate several critical brain regions as prototypes. Further, we introduce a contrastive loss function to constrain the prototypes distribution, which embed the hierarchical brain semantic structure into the latent space. Self-attention and GCN are then used to dynamically construct hierarchical correlations of critical regions for brain network and explore the correlation, respectively. Our method is evaluated on ADNI-1 and ADNI-2 databases for mild cognitive impairment (MCI) conversion prediction, and acheive the state-of-the-art (SOTA) performance. Our source code is available at http://github.com/*******.
Bilinear pooling and metric learning network for early Alzheimer's disease identification with FDG-PET images
Nov 09, 2021



Abstract:FDG-PET reveals altered brain metabolism in individuals with mild cognitive impairment (MCI) and Alzheimer's disease (AD). Some biomarkers derived from FDG-PET by computer-aided-diagnosis (CAD) technologies have been proved that they can accurately diagnosis normal control (NC), MCI, and AD. However, the studies of identification of early MCI (EMCI) and late MCI (LMCI) with FDG-PET images are still insufficient. Compared with studies based on fMRI and DTI images, the researches of the inter-region representation features in FDG-PET images are insufficient. Moreover, considering the variability in different individuals, some hard samples which are very similar with both two classes limit the classification performance. To tackle these problems, in this paper, we propose a novel bilinear pooling and metric learning network (BMNet), which can extract the inter-region representation features and distinguish hard samples by constructing embedding space. To validate the proposed method, we collect 998 FDG-PET images from ADNI. Following the common preprocessing steps, 90 features are extracted from each FDG-PET image according to the automatic anatomical landmark (AAL) template and then sent into the proposed network. Extensive 5-fold cross-validation experiments are performed for multiple two-class classifications. Experiments show that most metrics are improved after adding the bilinear pooling module and metric losses to the Baseline model respectively. Specifically, in the classification task between EMCI and LMCI, the specificity improves 6.38% after adding the triple metric loss, and the negative predictive value (NPV) improves 3.45% after using the bilinear pooling module.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge