Yaozhen Zhu
Explaining Graph Neural Networks with Large Language Models: A Counterfactual Perspective for Molecular Property Prediction
Oct 19, 2024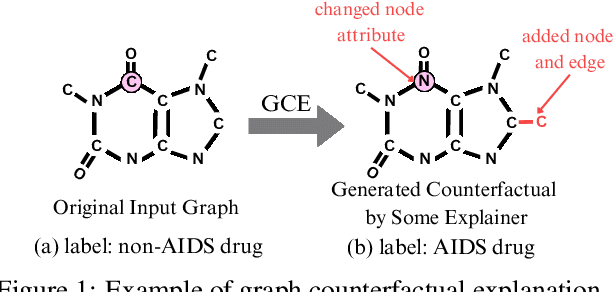



Abstract:In recent years, Graph Neural Networks (GNNs) have become successful in molecular property prediction tasks such as toxicity analysis. However, due to the black-box nature of GNNs, their outputs can be concerning in high-stakes decision-making scenarios, e.g., drug discovery. Facing such an issue, Graph Counterfactual Explanation (GCE) has emerged as a promising approach to improve GNN transparency. However, current GCE methods usually fail to take domain-specific knowledge into consideration, which can result in outputs that are not easily comprehensible by humans. To address this challenge, we propose a novel GCE method, LLM-GCE, to unleash the power of large language models (LLMs) in explaining GNNs for molecular property prediction. Specifically, we utilize an autoencoder to generate the counterfactual graph topology from a set of counterfactual text pairs (CTPs) based on an input graph. Meanwhile, we also incorporate a CTP dynamic feedback module to mitigate LLM hallucination, which provides intermediate feedback derived from the generated counterfactuals as an attempt to give more faithful guidance. Extensive experiments demonstrate the superior performance of LLM-GCE. Our code is released on https://github.com/YinhanHe123/new\_LLM4GNNExplanation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge