Wenxin Xue
PLLaMa: An Open-source Large Language Model for Plant Science
Jan 03, 2024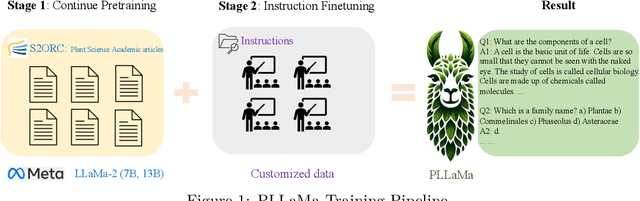

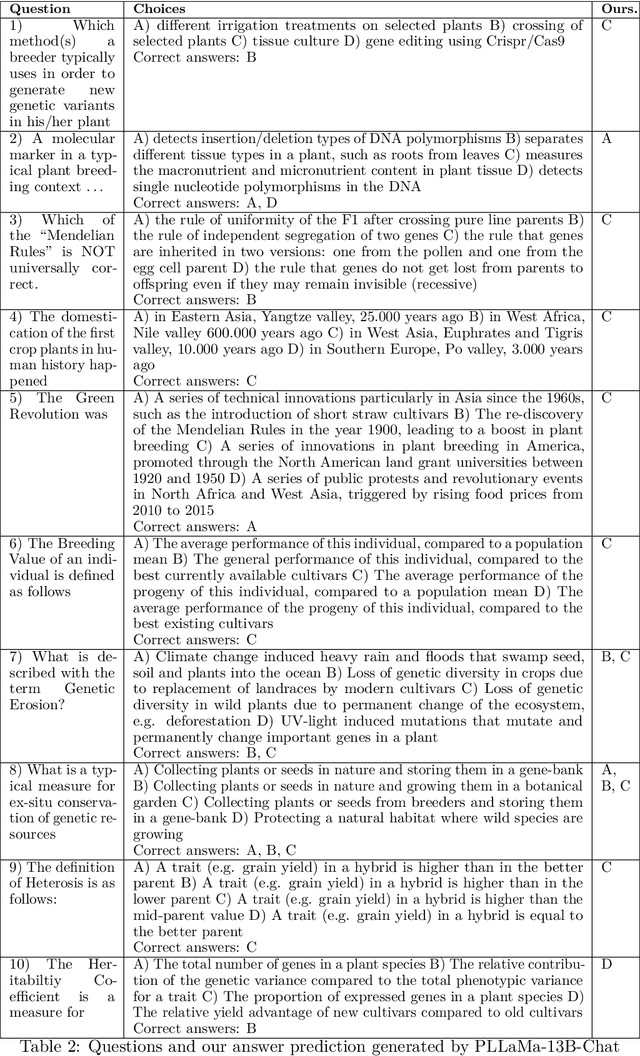
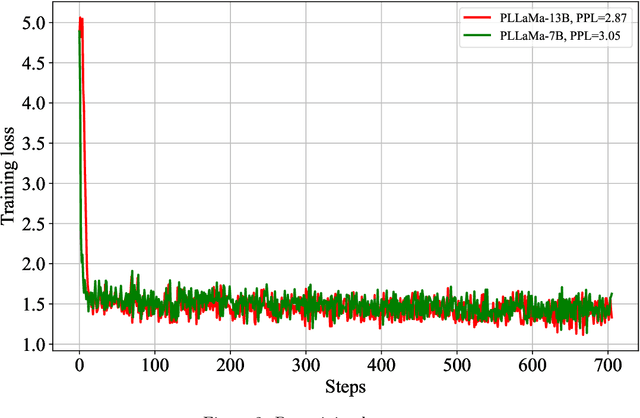
Abstract:Large Language Models (LLMs) have exhibited remarkable capabilities in understanding and interacting with natural language across various sectors. However, their effectiveness is limited in specialized areas requiring high accuracy, such as plant science, due to a lack of specific expertise in these fields. This paper introduces PLLaMa, an open-source language model that evolved from LLaMa-2. It's enhanced with a comprehensive database, comprising more than 1.5 million scholarly articles in plant science. This development significantly enriches PLLaMa with extensive knowledge and proficiency in plant and agricultural sciences. Our initial tests, involving specific datasets related to plants and agriculture, show that PLLaMa substantially improves its understanding of plant science-related topics. Moreover, we have formed an international panel of professionals, including plant scientists, agricultural engineers, and plant breeders. This team plays a crucial role in verifying the accuracy of PLLaMa's responses to various academic inquiries, ensuring its effective and reliable application in the field. To support further research and development, we have made the model's checkpoints and source codes accessible to the scientific community. These resources are available for download at \url{https://github.com/Xianjun-Yang/PLLaMa}.
Advancing Early Detection of Virus Yellows: Developing a Hybrid Convolutional Neural Network for Automatic Aphid Counting in Sugar Beet Fields
Aug 09, 2023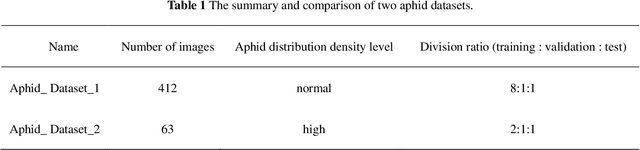
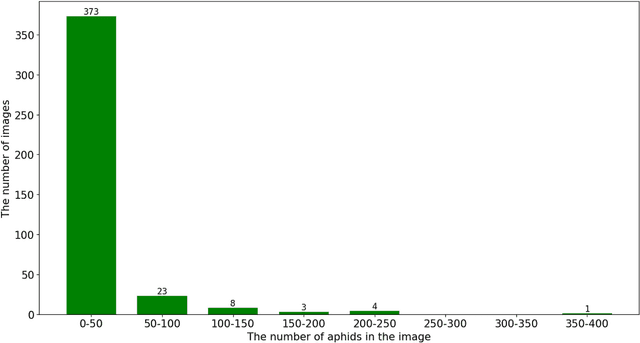
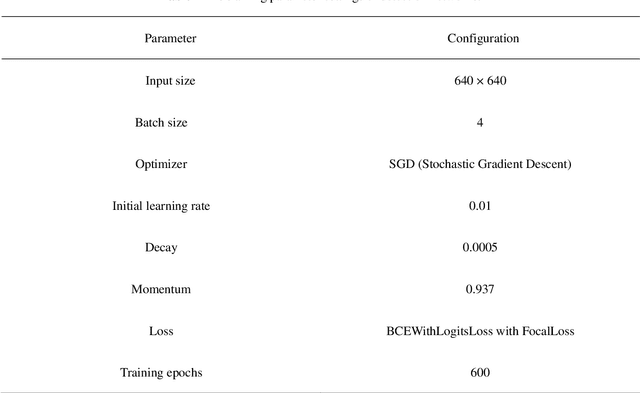
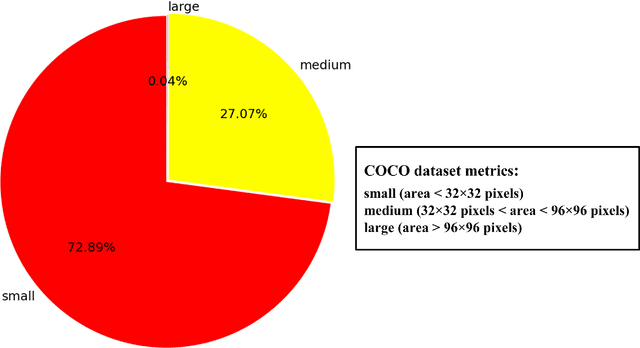
Abstract:Aphids are efficient vectors to transmit virus yellows in sugar beet fields. Timely monitoring and control of their populations are thus critical to prevent the large-scale outbreak of virus yellows. However, the manual counting of aphids, which is the most common practice, is labor-intensive and time-consuming. Additionally, two of the biggest challenges in aphid counting are that aphids are small objects and their density distributions are varied in different areas of the field. To address these challenges, we proposed a hybrid automatic aphid counting network architecture which integrates the detection network and the density map estimation network. When the distribution density of aphids is low, it utilizes an improved Yolov5 to count aphids. Conversely, when the distribution density of aphids is high, its witches to CSRNet to count aphids. To the best of our knowledge, this is the first framework integrating the detection network and the density map estimation network for counting tasks. Through comparison experiments of counting aphids, it verified that our proposed approach outperforms all other methods in counting aphids. It achieved the lowest MAE and RMSE values for both the standard and high-density aphid datasets: 2.93 and 4.01 (standard), and 34.19 and 38.66 (high-density), respectively. Moreover, the AP of the improved Yolov5 is 5% higher than that of the original Yolov5. Especially for extremely small aphids and densely distributed aphids, the detection performance of the improved Yolov5 is significantly better than the original Yolov5. This work provides an effective early warning for the virus yellows risk caused by aphids in sugar beet fields, offering protection for sugar beet growth and ensuring sugar beet yield. The datasets and project code are released at: https://github.com/JunfengGaolab/Counting-Aphids.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge