Weiyi Yu
FAN-Net: Fourier-Based Adaptive Normalization For Cross-Domain Stroke Lesion Segmentation
Apr 23, 2023



Abstract:Since stroke is the main cause of various cerebrovascular diseases, deep learning-based stroke lesion segmentation on magnetic resonance (MR) images has attracted considerable attention. However, the existing methods often neglect the domain shift among MR images collected from different sites, which has limited performance improvement. To address this problem, we intend to change style information without affecting high-level semantics via adaptively changing the low-frequency amplitude components of the Fourier transform so as to enhance model robustness to varying domains. Thus, we propose a novel FAN-Net, a U-Net--based segmentation network incorporated with a Fourier-based adaptive normalization (FAN) and a domain classifier with a gradient reversal layer. The FAN module is tailored for learning adaptive affine parameters for the amplitude components of different domains, which can dynamically normalize the style information of source images. Then, the domain classifier provides domain-agnostic knowledge to endow FAN with strong domain generalizability. The experimental results on the ATLAS dataset, which consists of MR images from 9 sites, show the superior performance of the proposed FAN-Net compared with baseline methods.
Site Generalization: Stroke Lesion Segmentation on Magnetic Resonance Images from Unseen Sites
May 09, 2022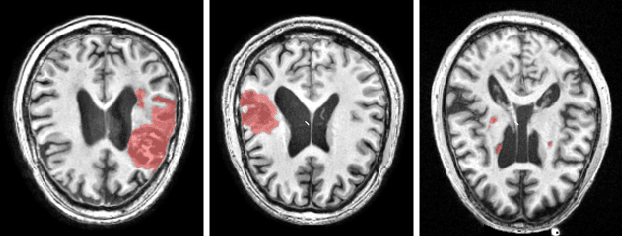
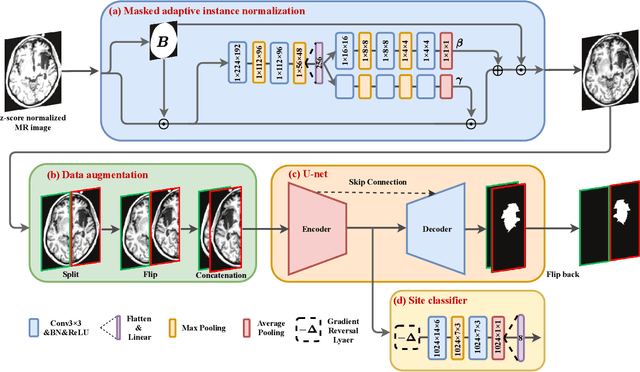
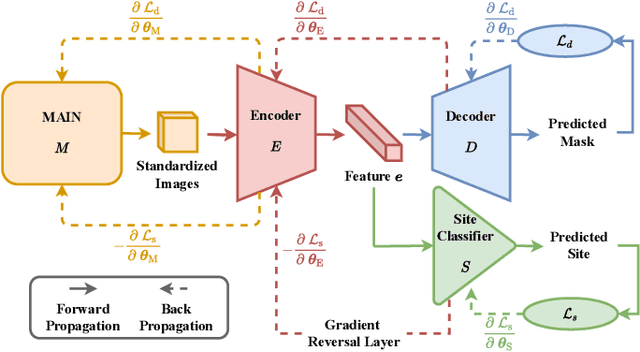
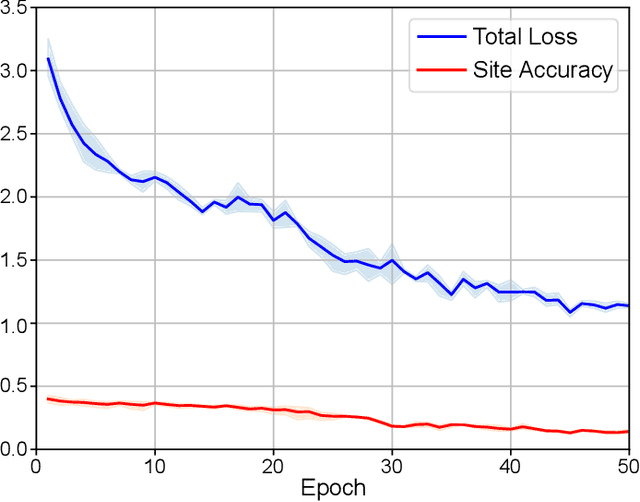
Abstract:There are considerable interests in automatic stroke lesion segmentation on magnetic resonance (MR) images in the medical imaging field, as strokes are the main cause of various cerebrovascular diseases. Although deep learning-based models have been proposed for this task, generalizing these models to unseen sites is difficult due to not only the large intersite discrepancy among different scanners, imaging protocols, and populations but also the variations in stroke lesion shape, size, and location. Thus, we propose a U-net--based segmentation network termed SG-Net to improve unseen site generalization for stroke lesion segmentation on MR images. Specifically, we first propose masked adaptive instance normalization (MAIN) to minimize intersite discrepancies, standardizing input MR images from different sites into a site-unrelated style by dynamically learning affine parameters from the input. Then, we leverage a gradient reversal layer to force the U-net encoder to learn site-invariant representation, which further improves the model generalization in conjunction with MAIN. Finally, inspired by the "pseudosymmetry" of the human brain, we introduce a simple, yet effective data augmentation technique that can be embedded within SG-Net to double the sample size while halving memory consumption. As a result, stroke lesions from the whole brain can be easily identified within a hemisphere, improving the simplicity of training. Experimental results on the benchmark Anatomical Tracings of Lesions After Stroke (ATLAS) dataset, which includes MR images from 9 different sites, demonstrate that under the "leave-one-site-out" setting, the proposed SG-Net substantially outperforms recently published methods in terms of quantitative metrics and qualitative comparisons.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge