Vishalini Laguduva Ramnath
Metagenome2Vec: Building Contextualized Representations for Scalable Metagenome Analysis
Nov 09, 2021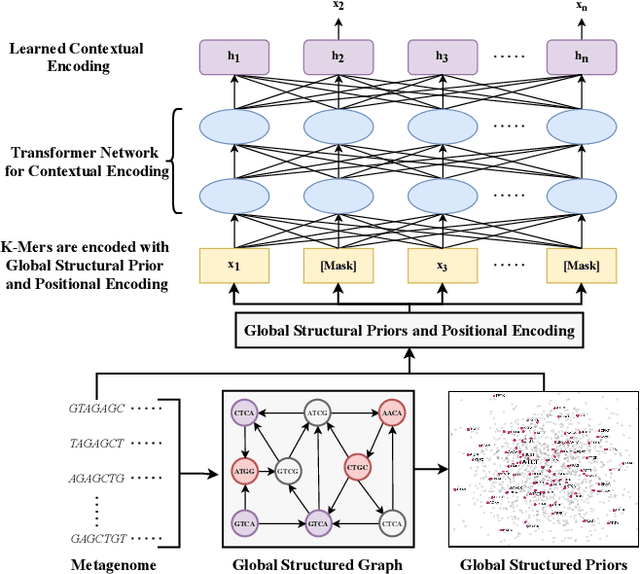
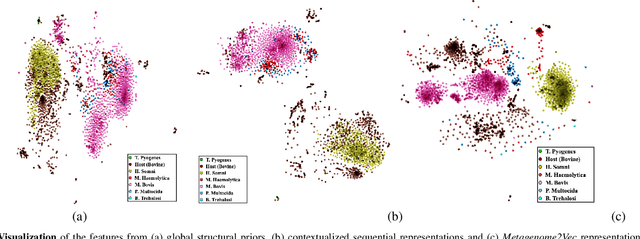
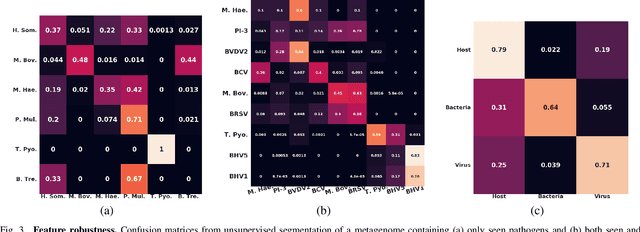
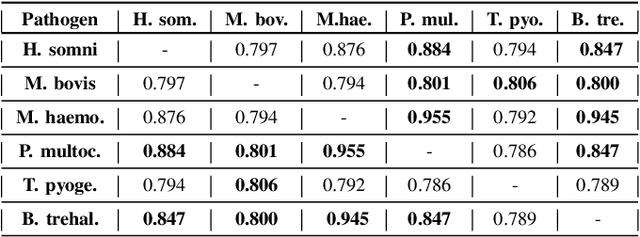
Abstract:Advances in next-generation metagenome sequencing have the potential to revolutionize the point-of-care diagnosis of novel pathogen infections, which could help prevent potential widespread transmission of diseases. Given the high volume of metagenome sequences, there is a need for scalable frameworks to analyze and segment metagenome sequences from clinical samples, which can be highly imbalanced. There is an increased need for learning robust representations from metagenome reads since pathogens within a family can have highly similar genome structures (some more than 90%) and hence enable the segmentation and identification of novel pathogen sequences with limited labeled data. In this work, we propose Metagenome2Vec - a contextualized representation that captures the global structural properties inherent in metagenome data and local contextualized properties through self-supervised representation learning. We show that the learned representations can help detect six (6) related pathogens from clinical samples with less than 100 labeled sequences. Extensive experiments on simulated and clinical metagenome data show that the proposed representation encodes compositional properties that can generalize beyond annotations to segment novel pathogens in an unsupervised setting.
MG-NET: Leveraging Pseudo-Imaging for Multi-Modal Metagenome Analysis
Jul 21, 2021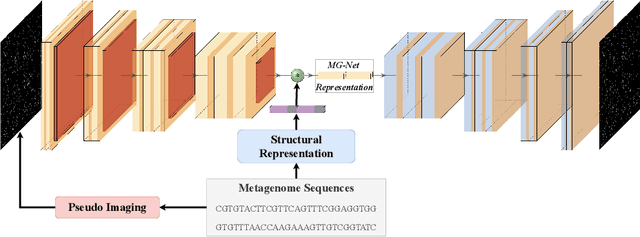
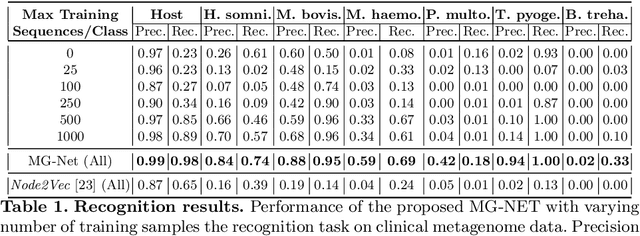
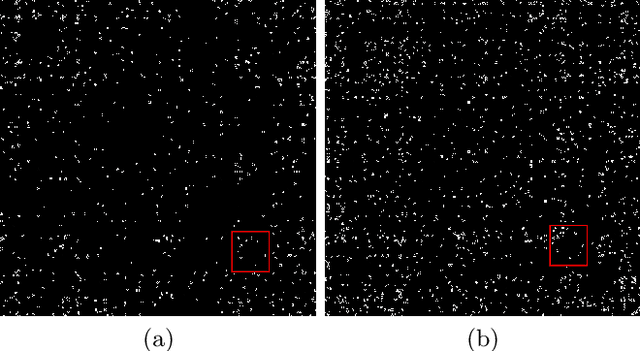
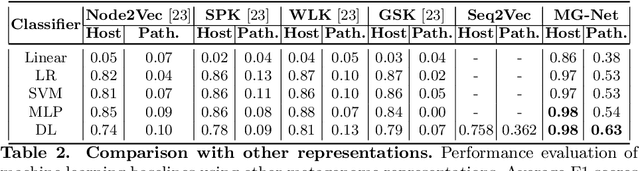
Abstract:The emergence of novel pathogens and zoonotic diseases like the SARS-CoV-2 have underlined the need for developing novel diagnosis and intervention pipelines that can learn rapidly from small amounts of labeled data. Combined with technological advances in next-generation sequencing, metagenome-based diagnostic tools hold much promise to revolutionize rapid point-of-care diagnosis. However, there are significant challenges in developing such an approach, the chief among which is to learn self-supervised representations that can help detect novel pathogen signatures with very low amounts of labeled data. This is particularly a difficult task given that closely related pathogens can share more than 90% of their genome structure. In this work, we address these challenges by proposing MG-Net, a self-supervised representation learning framework that leverages multi-modal context using pseudo-imaging data derived from clinical metagenome sequences. We show that the proposed framework can learn robust representations from unlabeled data that can be used for downstream tasks such as metagenome sequence classification with limited access to labeled data. Extensive experiments show that the learned features outperform current baseline metagenome representations, given only 1000 samples per class.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge