Vincent De Andrade
Murine AI excels at cats and cheese: Structural differences between human and mouse neurons and their implementation in generative AIs
Oct 28, 2024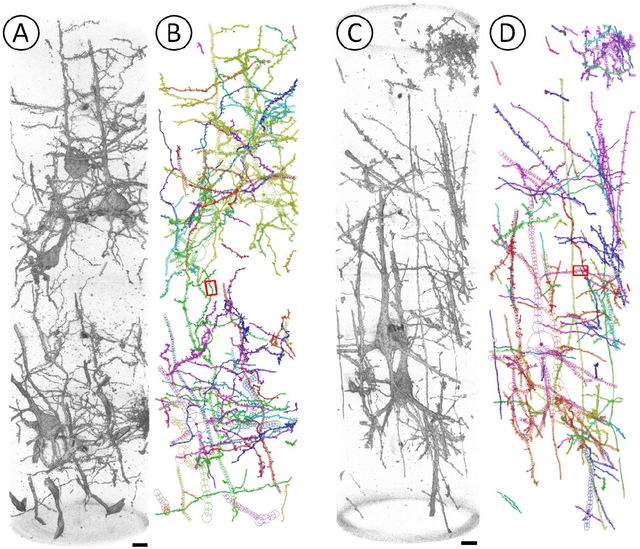
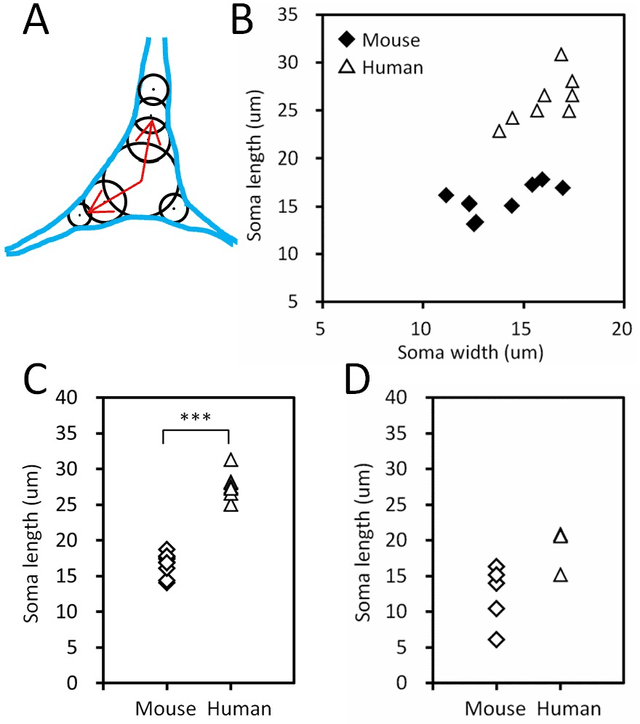
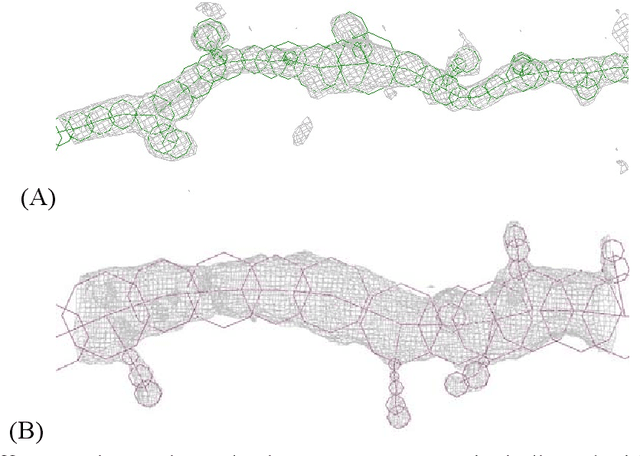
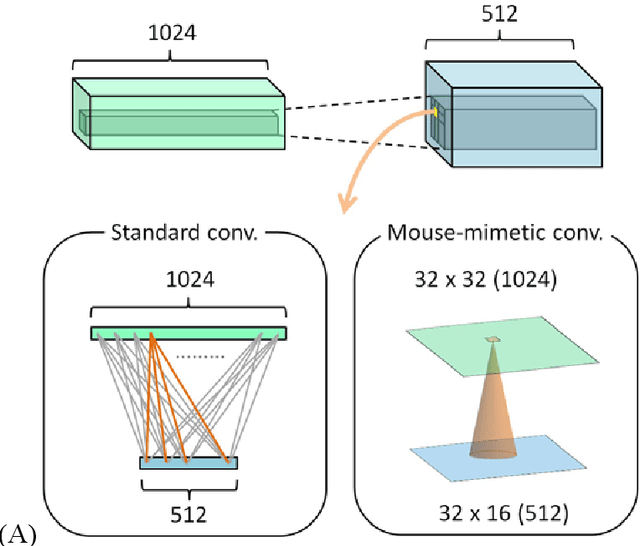
Abstract:Mouse and human brains have different functions that depend on their neuronal networks. In this study, we analyzed nanometer-scale three-dimensional structures of brain tissues of the mouse medial prefrontal cortex and compared them with structures of the human anterior cingulate cortex. The obtained results indicated that mouse neuronal somata are smaller and neurites are thinner than those of human neurons. These structural features allow mouse neurons to be integrated in the limited space of the brain, though thin neurites should suppress distal connections according to cable theory. We implemented this mouse-mimetic constraint in convolutional layers of a generative adversarial network (GAN) and a denoising diffusion implicit model (DDIM), which were then subjected to image generation tasks using photo datasets of cat faces, cheese, human faces, and birds. The mouse-mimetic GAN outperformed a standard GAN in the image generation task using the cat faces and cheese photo datasets, but underperformed for human faces and birds. The mouse-mimetic DDIM gave similar results, suggesting that the nature of the datasets affected the results. Analyses of the four datasets indicated differences in their image entropy, which should influence the number of parameters required for image generation. The preferences of the mouse-mimetic AIs coincided with the impressions commonly associated with mice. The relationship between the neuronal network and brain function should be investigated by implementing other biological findings in artificial neural networks.
Adorym: A multi-platform generic x-ray image reconstruction framework based on automatic differentiation
Dec 22, 2020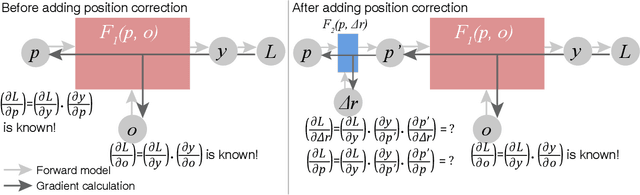
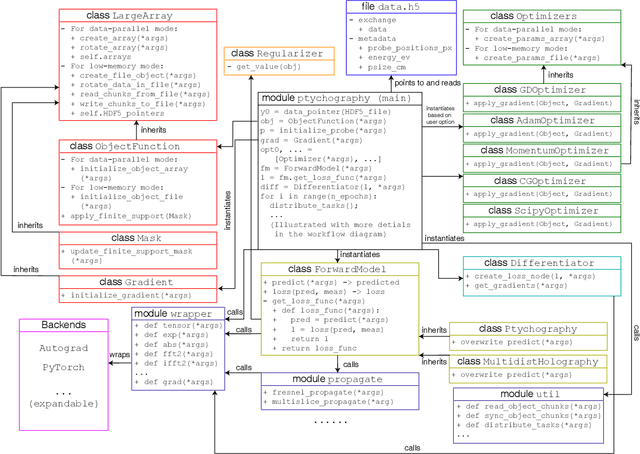
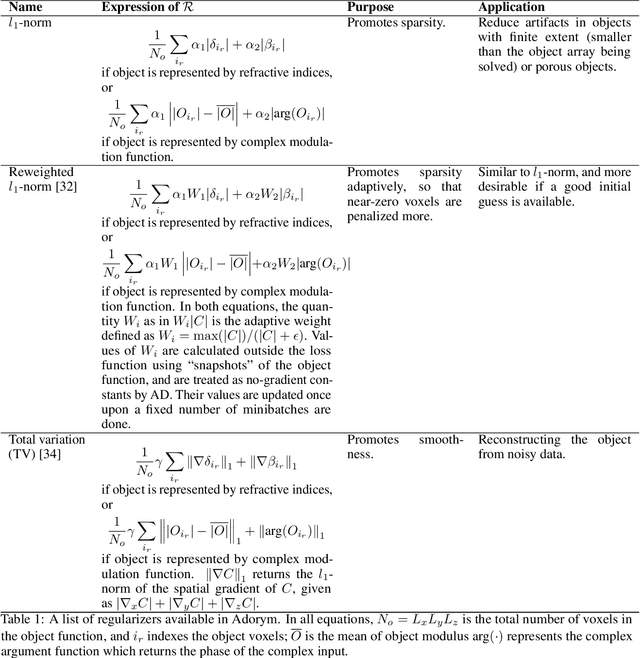
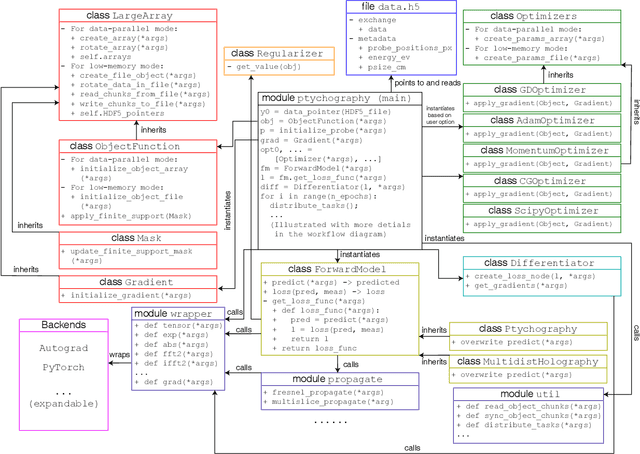
Abstract:We describe and demonstrate an optimization-based x-ray image reconstruction framework called Adorym. Our framework provides a generic forward model, allowing one code framework to be used for a wide range of imaging methods ranging from near-field holography to and fly-scan ptychographic tomography. By using automatic differentiation for optimization, Adorym has the flexibility to refine experimental parameters including probe positions, multiple hologram alignment, and object tilts. It is written with strong support for parallel processing, allowing large datasets to be processed on high-performance computing systems. We demonstrate its use on several experimental datasets to show improved image quality through parameter refinement.
Quantifying mesoscale neuroanatomy using X-ray microtomography
Jul 26, 2016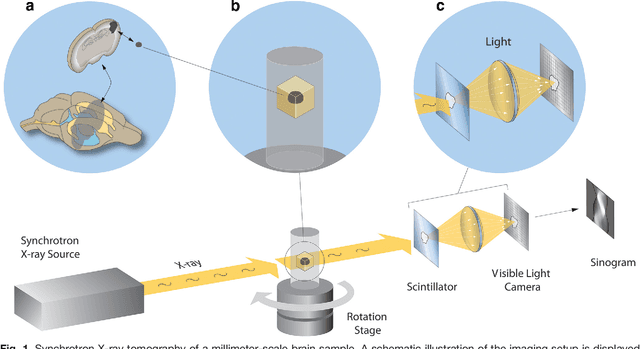
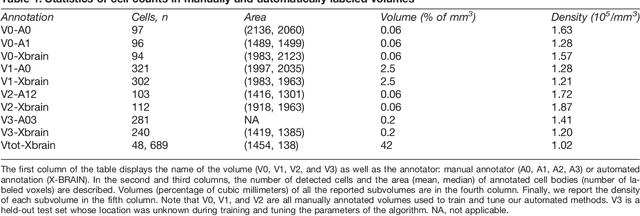
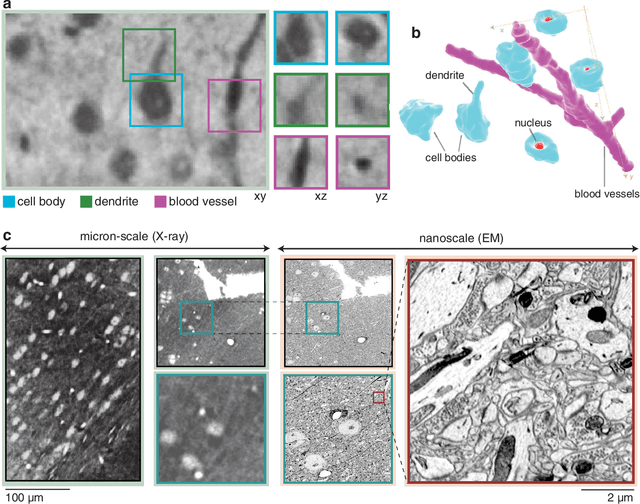
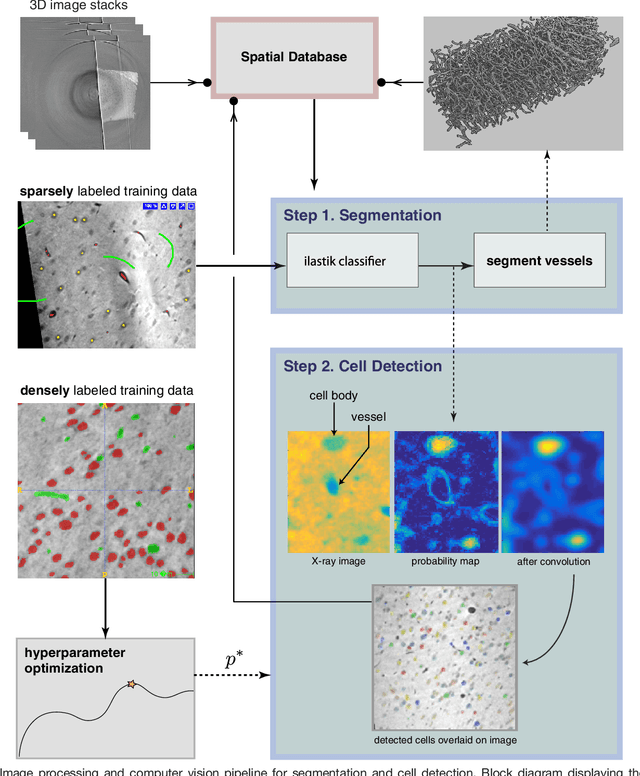
Abstract:Methods for resolving the 3D microstructure of the brain typically start by thinly slicing and staining the brain, and then imaging each individual section with visible light photons or electrons. In contrast, X-rays can be used to image thick samples, providing a rapid approach for producing large 3D brain maps without sectioning. Here we demonstrate the use of synchrotron X-ray microtomography ($\mu$CT) for producing mesoscale $(1~\mu m^3)$ resolution brain maps from millimeter-scale volumes of mouse brain. We introduce a pipeline for $\mu$CT-based brain mapping that combines methods for sample preparation, imaging, automated segmentation of image volumes into cells and blood vessels, and statistical analysis of the resulting brain structures. Our results demonstrate that X-ray tomography promises rapid quantification of large brain volumes, complementing other brain mapping and connectomics efforts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge