Vikram Nathan
Tsunami: A Learned Multi-dimensional Index for Correlated Data and Skewed Workloads
Jun 23, 2020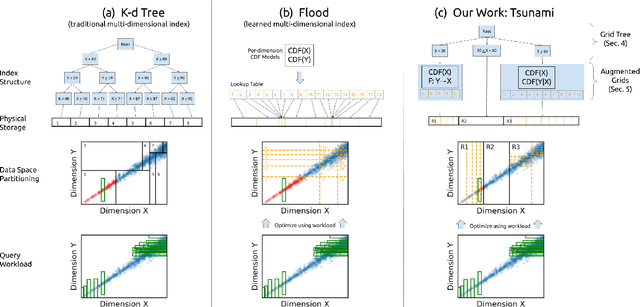

Abstract:Filtering data based on predicates is one of the most fundamental operations for any modern data warehouse. Techniques to accelerate the execution of filter expressions include clustered indexes, specialized sort orders (e.g., Z-order), multi-dimensional indexes, and, for high selectivity queries, secondary indexes. However, these schemes are hard to tune and their performance is inconsistent. Recent work on learned multi-dimensional indexes has introduced the idea of automatically optimizing an index for a particular dataset and workload. However, the performance of that work suffers in the presence of correlated data and skewed query workloads, both of which are common in real applications. In this paper, we introduce Tsunami, which addresses these limitations to achieve up to 6X faster query performance and up to 8X smaller index size than existing learned multi-dimensional indexes, in addition to up to 11X faster query performance and 170X smaller index size than optimally-tuned traditional indexes.
Learning Multi-dimensional Indexes
Dec 03, 2019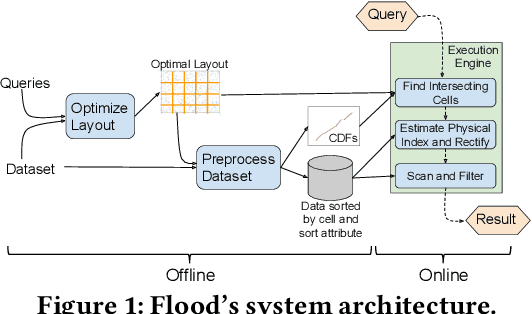
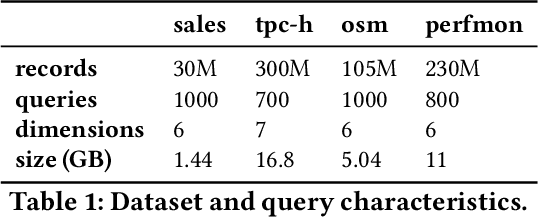


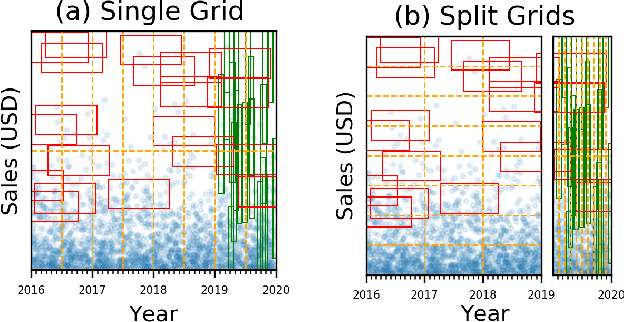
Abstract:Scanning and filtering over multi-dimensional tables are key operations in modern analytical database engines. To optimize the performance of these operations, databases often create clustered indexes over a single dimension or multi-dimensional indexes such as R-trees, or use complex sort orders (e.g., Z-ordering). However, these schemes are often hard to tune and their performance is inconsistent across different datasets and queries. In this paper, we introduce Flood, a multi-dimensional in-memory index that automatically adapts itself to a particular dataset and workload by jointly optimizing the index structure and data storage. Flood achieves up to three orders of magnitude faster performance for range scans with predicates than state-of-the-art multi-dimensional indexes or sort orders on real-world datasets and workloads. Our work serves as a building block towards an end-to-end learned database system.
LISA: Towards Learned DNA Sequence Search
Oct 10, 2019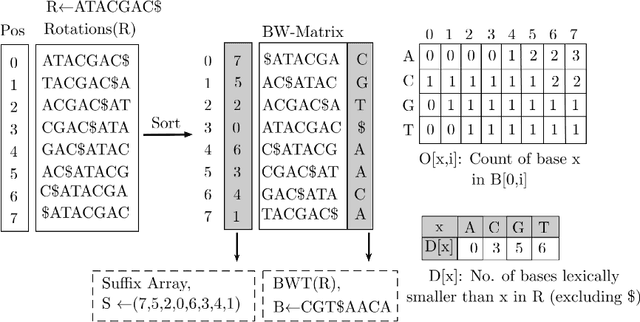

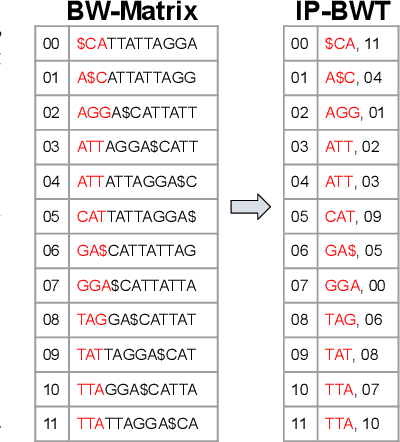
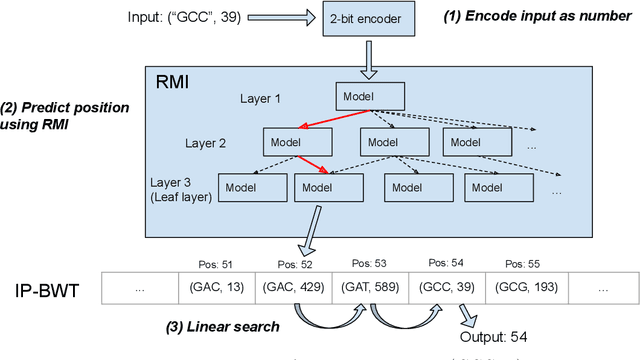
Abstract:Next-generation sequencing (NGS) technologies have enabled affordable sequencing of billions of short DNA fragments at high throughput, paving the way for population-scale genomics. Genomics data analytics at this scale requires overcoming performance bottlenecks, such as searching for short DNA sequences over long reference sequences. In this paper, we introduce LISA (Learned Indexes for Sequence Analysis), a novel learning-based approach to DNA sequence search. As a first proof of concept, we focus on accelerating one of the most essential flavors of the problem, called exact search. LISA builds on and extends FM-index, which is the state-of-the-art technique widely deployed in genomics tool-chains. Initial experiments with human genome datasets indicate that LISA achieves up to a factor of 4X performance speedup against its traditional counterpart.
Accurate Streaming Support Vector Machines
Dec 08, 2014


Abstract:A widely-used tool for binary classification is the Support Vector Machine (SVM), a supervised learning technique that finds the "maximum margin" linear separator between the two classes. While SVMs have been well studied in the batch (offline) setting, there is considerably less work on the streaming (online) setting, which requires only a single pass over the data using sub-linear space. Existing streaming algorithms are not yet competitive with the batch implementation. In this paper, we use the formulation of the SVM as a minimum enclosing ball (MEB) problem to provide a streaming SVM algorithm based off of the blurred ball cover originally proposed by Agarwal and Sharathkumar. Our implementation consistently outperforms existing streaming SVM approaches and provides higher accuracies than libSVM on several datasets, thus making it competitive with the standard SVM batch implementation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge