Vidya Ganapati
Self-Supervised Deep Learning for Model Correction in the Computational Crystallography Toolbox
Jul 04, 2023Abstract:The Computational Crystallography Toolbox (CCTBX) is open-source software that allows for processing of crystallographic data, including from serial femtosecond crystallography (SFX), for macromolecular structure determination. We aim to use the modules in CCTBX to determine the oxidation state of individual metal atoms in a macromolecule. Changes in oxidation state are reflected in small shifts of the atom's X-ray absorption edge. These energy shifts can be extracted from the diffraction images recorded in serial femtosecond crystallography, given knowledge of a forward physics model. However, as the diffraction changes only slightly due to the absorption edge shift, inaccuracies in the forward physics model make it extremely challenging to observe the oxidation state. In this work, we describe the potential impact of using self-supervised deep learning to correct the scientific model in CCTBX and provide uncertainty quantification. We provide code for forward model simulation and data analysis, built from CCTBX modules, at https://github.com/gigantocypris/SPREAD , which can be integrated with machine learning. We describe open questions in algorithm development to help spur advances through dialog between crystallographers and machine learning researchers. New methods could help elucidate charge transfer processes in many reactions, including key events in photosynthesis.
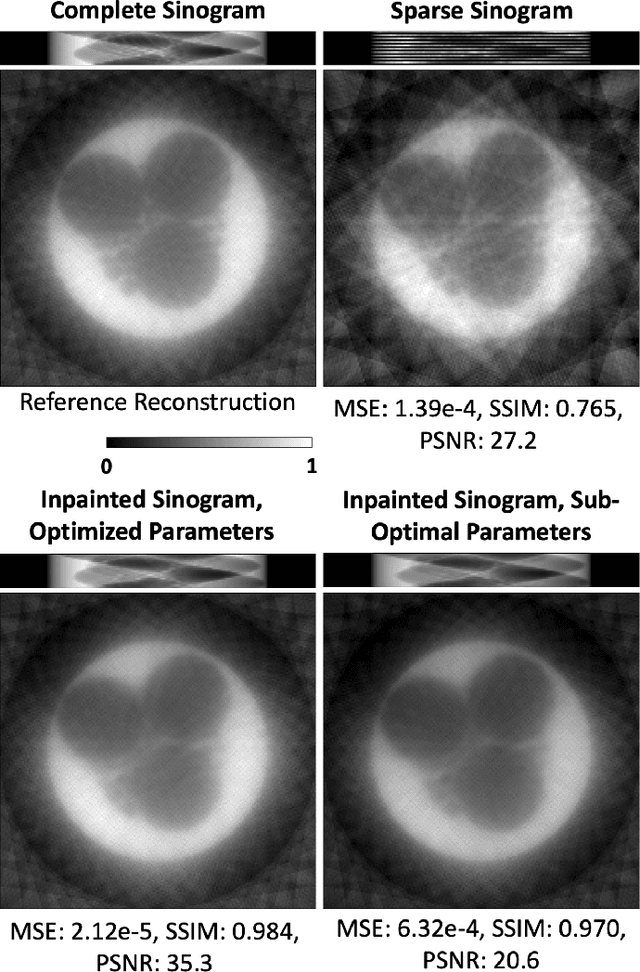
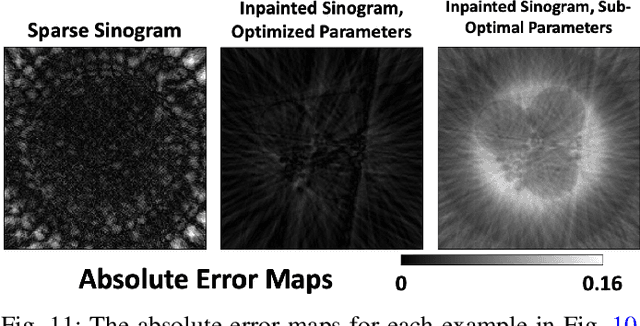
A Self-Supervised Approach to Reconstruction in Sparse X-Ray Computed Tomography
Oct 30, 2022Abstract:Computed tomography has propelled scientific advances in fields from biology to materials science. This technology allows for the elucidation of 3-dimensional internal structure by the attenuation of x-rays through an object at different rotations relative to the beam. By imaging 2-dimensional projections, a 3-dimensional object can be reconstructed through a computational algorithm. Imaging at a greater number of rotation angles allows for improved reconstruction. However, taking more measurements increases the x-ray dose and may cause sample damage. Deep neural networks have been used to transform sparse 2-D projection measurements to a 3-D reconstruction by training on a dataset of known similar objects. However, obtaining high-quality object reconstructions for the training dataset requires high x-ray dose measurements that can destroy or alter the specimen before imaging is complete. This becomes a chicken-and-egg problem: high-quality reconstructions cannot be generated without deep learning, and the deep neural network cannot be learned without the reconstructions. This work develops and validates a self-supervised probabilistic deep learning technique, the physics-informed variational autoencoder, to solve this problem. A dataset consisting solely of sparse projection measurements from each object is used to jointly reconstruct all objects of the set. This approach has the potential to allow visualization of fragile samples with x-ray computed tomography. We release our code for reproducing our results at: https://github.com/vganapati/CT_PVAE .
Data-Driven Computational Imaging for Scientific Discovery
Oct 29, 2022Abstract:In computational imaging, hardware for signal sampling and software for object reconstruction are designed in tandem for improved capability. Examples of such systems include computed tomography (CT), magnetic resonance imaging (MRI), and superresolution microscopy. In contrast to more traditional cameras, in these devices, indirect measurements are taken and computational algorithms are used for reconstruction. This allows for advanced capabilities such as super-resolution or 3-dimensional imaging, pushing forward the frontier of scientific discovery. However, these techniques generally require a large number of measurements, causing low throughput, motion artifacts, and/or radiation damage, limiting applications. Data-driven approaches to reducing the number of measurements needed have been proposed, but they predominately require a ground truth or reference dataset, which may be impossible to collect. This work outlines a self-supervised approach and explores the future work that is necessary to make such a technique usable for real applications. Light-emitting diode (LED) array microscopy, a modality that allows visualization of transparent objects in two and three dimensions with high resolution and field-of-view, is used as an illustrative example. We release our code at https://github.com/vganapati/LED_PVAE and our experimental data at https://doi.org/10.6084/m9.figshare.21232088 .
HYPPO: A Surrogate-Based Multi-Level Parallelism Tool for Hyperparameter Optimization
Oct 04, 2021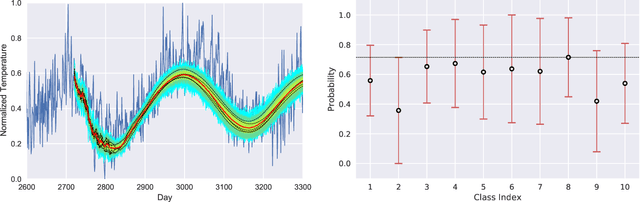
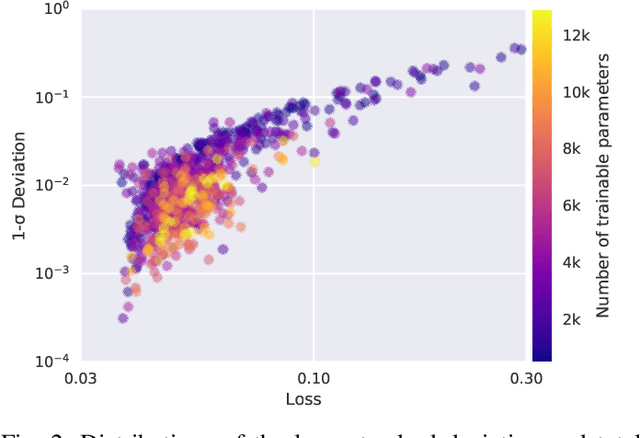
Abstract:We present a new software, HYPPO, that enables the automatic tuning of hyperparameters of various deep learning (DL) models. Unlike other hyperparameter optimization (HPO) methods, HYPPO uses adaptive surrogate models and directly accounts for uncertainty in model predictions to find accurate and reliable models that make robust predictions. Using asynchronous nested parallelism, we are able to significantly alleviate the computational burden of training complex architectures and quantifying the uncertainty. HYPPO is implemented in Python and can be used with both TensorFlow and PyTorch libraries. We demonstrate various software features on time-series prediction and image classification problems as well as a scientific application in computed tomography image reconstruction. Finally, we show that (1) we can reduce by an order of magnitude the number of evaluations necessary to find the most optimal region in the hyperparameter space and (2) we can reduce by two orders of magnitude the throughput for such HPO process to complete.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge