Upul Senanayake
Information Transfer in Swarms with Leaders
Jun 30, 2014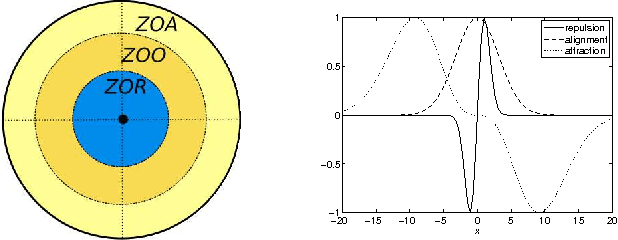
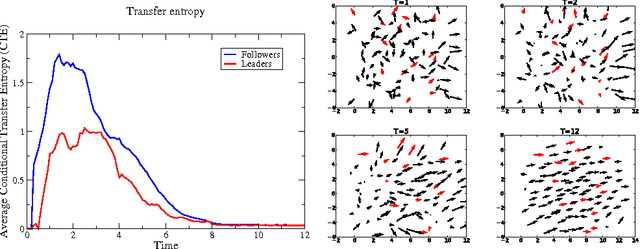
Abstract:Swarm dynamics is the study of collections of agents that interact with one another without central control. In natural systems, insects, birds, fish and other large mammals function in larger units to increase the overall fitness of the individuals. Their behavior is coordinated through local interactions to enhance mate selection, predator detection, migratory route identification and so forth [Andersson and Wallander 2003; Buhl et al. 2006; Nagy et al. 2010; Partridge 1982; Sumpter et al. 2008]. In artificial systems, swarms of autonomous agents can augment human activities such as search and rescue, and environmental monitoring by covering large areas with multiple nodes [Alami et al. 2007; Caruso et al. 2008; Ogren et al. 2004; Paley et al. 2007; Sibley et al. 2002]. In this paper, we explore the interplay between swarm dynamics, covert leadership and theoretical information transfer. A leader is a member of the swarm that acts upon information in addition to what is provided by local interactions. Depending upon the leadership model, leaders can use their external information either all the time or in response to local conditions [Couzin et al. 2005; Sun et al. 2013]. A covert leader is a leader that is treated no differently than others in the swarm, so leaders and followers participate equally in whatever interaction model is used [Rossi et al. 2007]. In this study, we use theoretical information transfer as a means of analyzing swarm interactions to explore whether or not it is possible to distinguish between followers and leaders based on interactions within the swarm. We find that covert leaders can be distinguished from followers in a swarm because they receive less transfer entropy than followers.
High Throughput Virtual Screening with Data Level Parallelism in Multi-core Processors
Dec 04, 2013
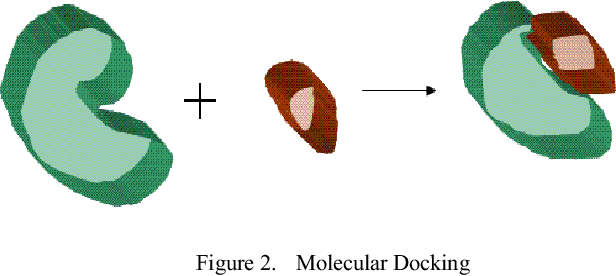
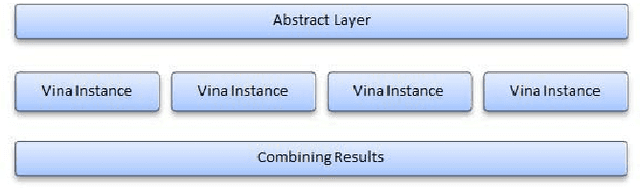
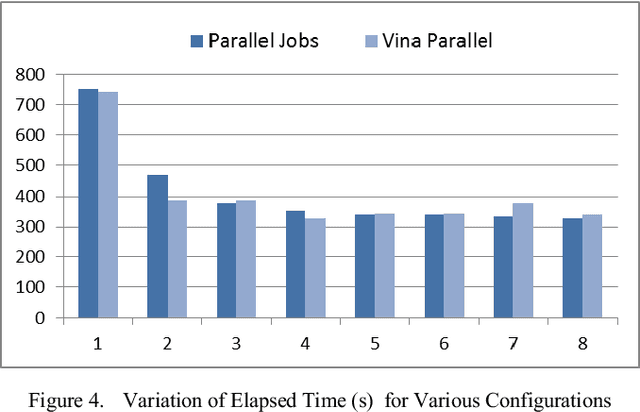
Abstract:Improving the throughput of molecular docking, a computationally intensive phase of the virtual screening process, is a highly sought area of research since it has a significant weight in the drug designing process. With such improvements, the world might find cures for incurable diseases like HIV disease and Cancer sooner. Our approach presented in this paper is to utilize a multi-core environment to introduce Data Level Parallelism (DLP) to the Autodock Vina software, which is a widely used for molecular docking software. Autodock Vina already exploits Instruction Level Parallelism (ILP) in multi-core environments and therefore optimized for such environments. However, with the results we have obtained, it can be clearly seen that our approach has enhanced the throughput of the already optimized software by more than six times. This will dramatically reduce the time consumed for the lead identification phase in drug designing along with the shift in the processor technology from multi-core to many-core of the current era. Therefore, we believe that the contribution of this project will effectively make it possible to expand the number of small molecules docked against a drug target and improving the chances to design drugs for incurable diseases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge