Toshiyo Tamura
Enhancement on Model Interpretability and Sleep Stage Scoring Performance with A Novel Pipeline Based on Deep Neural Network
Apr 07, 2022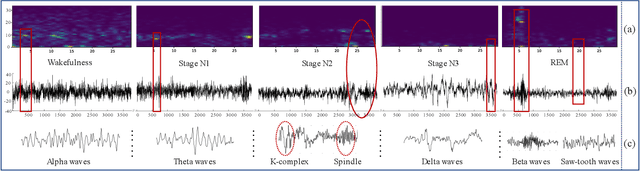
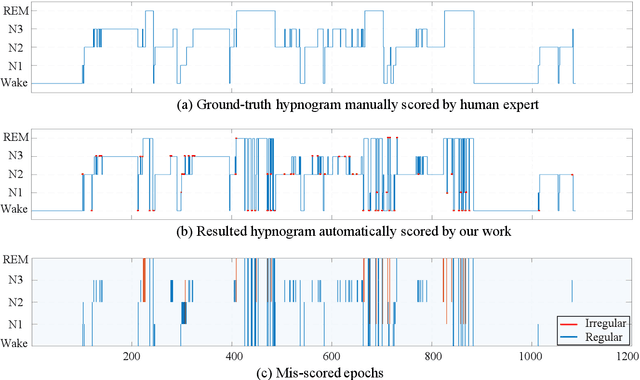
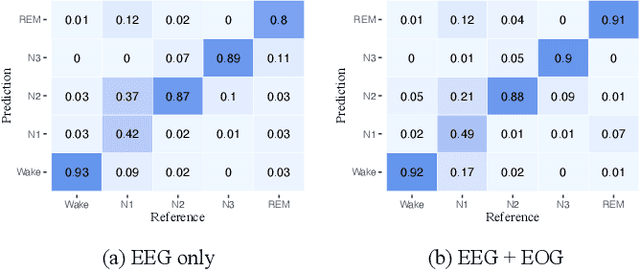
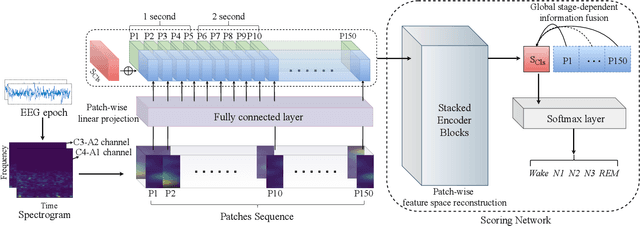
Abstract:Considering the natural frequency characteristics in sleep medicine, this paper first proposes a time-frequency framework for the representation learning of the electroencephalogram (EEG) following the definition of the American Academy of Sleep Medicine. To meet the temporal-random and transient nature of the defining characteristics of sleep stages, we further design a context-sensitive flexible pipeline that automatically adapts to the attributes of data itself. That is, the input EEG spectrogram is partitioned into a sequence of patches in the time and frequency axes, and then input to a delicate deep learning network for further representation learning to extract the stage-dependent features, which are used in the classification step finally. The proposed pipeline is validated against a large database, i.e., the Sleep Heart Health Study (SHHS), and the results demonstrate that the competitive performance for the wake, N2, and N3 stages outperforms the state-of-art works, with the F1 scores being 0.93, 0.88, and 0.87, respectively, and the proposed method has a high inter-rater reliability of 0.80 kappa. Importantly, we visualize the stage scoring process of the model decision with the Layer-wise Relevance Propagation (LRP) method, which shows that the proposed pipeline is more sensitive and perceivable in the decision-making process than the baseline pipelines. Therefore, the pipeline together with the LRP method can provide better model interpretability, which is important for clinical support.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge