Tom Michoel
Causal inference in drug discovery and development
Sep 29, 2022



Abstract:To discover new drugs is to seek and to prove causality. As an emerging approach leveraging human knowledge and creativity, data, and machine intelligence, causal inference holds the promise of reducing cognitive bias and improving decision making in drug discovery. While it has been applied across the value chain, the concepts and practice of causal inference remain obscure to many practitioners. This article offers a non-technical introduction to causal inference, reviews its recent applications, and discusses opportunities and challenges of adopting the causal language in drug discovery and development.
rfPhen2Gen: A machine learning based association study of brain imaging phenotypes to genotypes
Mar 31, 2022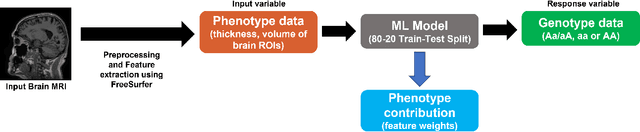



Abstract:Imaging genetic studies aim to find associations between genetic variants and imaging quantitative traits. Traditional genome-wide association studies (GWAS) are based on univariate statistical tests, but when multiple traits are analyzed together they suffer from a multiple-testing problem and from not taking into account correlations among the traits. An alternative approach to multi-trait GWAS is to reverse the functional relation between genotypes and traits, by fitting a multivariate regression model to predict genotypes from multiple traits simultaneously. However, current reverse genotype prediction approaches are mostly based on linear models. Here, we evaluated random forest regression (RFR) as a method to predict SNPs from imaging QTs and identify biologically relevant associations. We learned machine learning models to predict 518,484 SNPs using 56 brain imaging QTs. We observed that genotype regression error is a better indicator of permutation p-value significance than genotype classification accuracy. SNPs within the known Alzheimer disease (AD) risk gene APOE had lowest RMSE for lasso and random forest, but not ridge regression. Moreover, random forests identified additional SNPs that were not prioritized by the linear models but are known to be associated with brain-related disorders. Feature selection identified well-known brain regions associated with AD,like the hippocampus and amygdala, as important predictors of the most significant SNPs. In summary, our results indicate that non-linear methods like random forests may offer additional insights into phenotype-genotype associations compared to traditional linear multi-variate GWAS methods.
High-dimensional multi-trait GWAS by reverse prediction of genotypes
Oct 29, 2021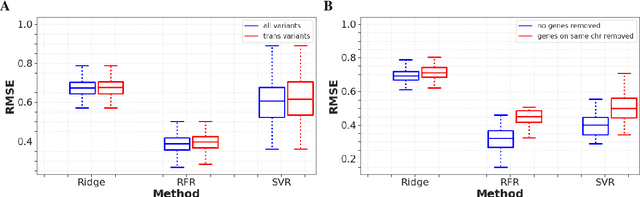
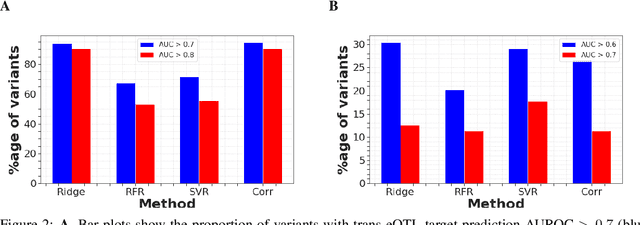
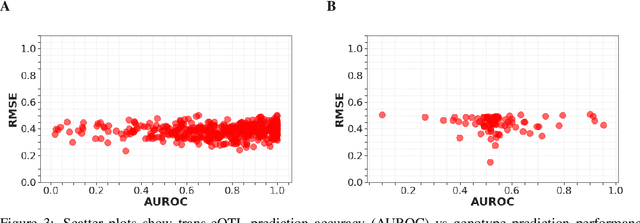
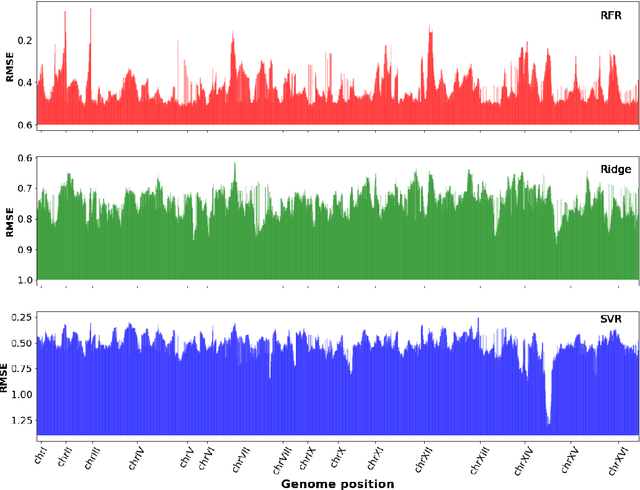
Abstract:Multi-trait genome-wide association studies (GWAS) use multi-variate statistical methods to identify associations between genetic variants and multiple correlated traits simultaneously, and have higher statistical power than independent univariate analysis of traits. Reverse regression, where genotypes of genetic variants are regressed on multiple traits simultaneously, has emerged as a promising approach to perform multi-trait GWAS in high-dimensional settings where the number of traits exceeds the number of samples. We extended this approach and analyzed different machine learning methods (ridge regression, random forests and support vector machines)for reverse regression in multi-trait GWAS, using genotypes, gene expression data and ground-truth transcriptional regulatory networks from the DREAM5 SysGen Challenge and from a cross between two yeast strains to evaluate methods. We found that genotype prediction performance, in terms of root mean squared error (RMSE), allowed to distinguish between genomic regions with high and low transcriptional activity. Moreover, model feature coefficients correlated with the strength of association between variants and individual traits, and were predictive of true trans-eQTL target genes, with complementary findings across methods.
Integrating Sensing and Communication in Cellular Networks via NR Sidelink
Sep 15, 2021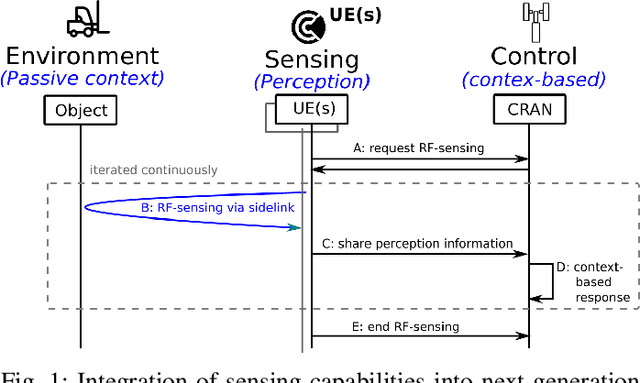

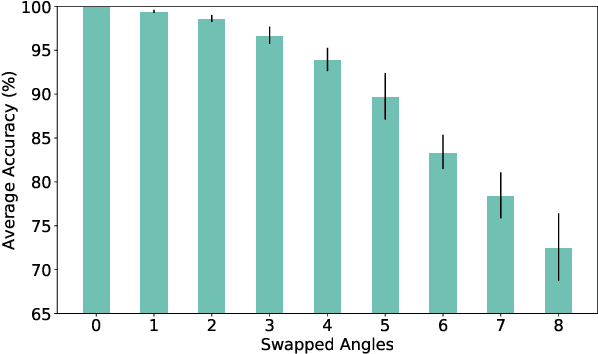
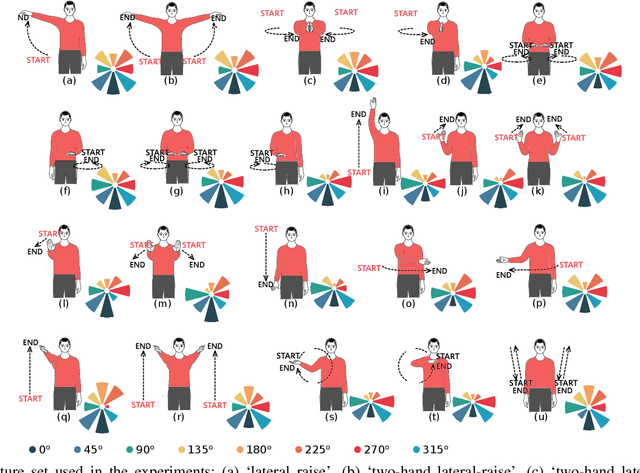
Abstract:RF-sensing, the analysis and interpretation of movement or environment-induced patterns in received electromagnetic signals, has been actively investigated for more than a decade. Since electromagnetic signals, through cellular communication systems, are omnipresent, RF sensing has the potential to become a universal sensing mechanism with applications in smart home, retail, localization, gesture recognition, intrusion detection, etc. Specifically, existing cellular network installations might be dual-used for both communication and sensing. Such communications and sensing convergence is envisioned for future communication networks. We propose the use of NR-sidelink direct device-to-device communication to achieve device-initiated,flexible sensing capabilities in beyond 5G cellular communication systems. In this article, we specifically investigate a common issue related to sidelink-based RF-sensing, which is its angle and rotation dependence. In particular, we discuss transformations of mmWave point-cloud data which achieve rotational invariance, as well as distributed processing based on such rotational invariant inputs, at angle and distance diverse devices. To process the distributed data, we propose a graph based encoder to capture spatio-temporal features of the data and propose four approaches for multi-angle learning. The approaches are compared on a newly recorded and openly available dataset comprising 15 subjects, performing 21 gestures which are recorded from 8 angles.
Tesla-Rapture: A Lightweight Gesture Recognition System from mmWave Radar Point Clouds
Sep 14, 2021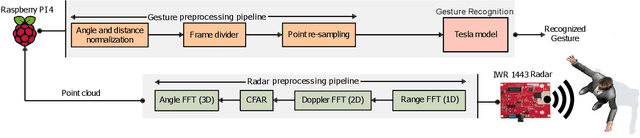
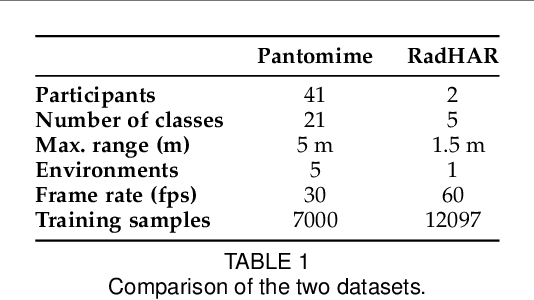

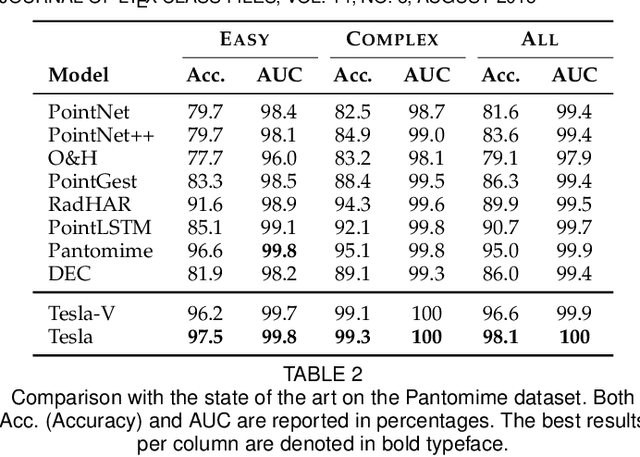
Abstract:We present Tesla-Rapture, a gesture recognition interface for point clouds generated by mmWave Radars. State of the art gesture recognition models are either too resource consuming or not sufficiently accurate for integration into real-life scenarios using wearable or constrained equipment such as IoT devices (e.g. Raspberry PI), XR hardware (e.g. HoloLens), or smart-phones. To tackle this issue, we developed Tesla, a Message Passing Neural Network (MPNN) graph convolution approach for mmWave radar point clouds. The model outperforms the state of the art on two datasets in terms of accuracy while reducing the computational complexity and, hence, the execution time. In particular, the approach, is able to predict a gesture almost 8 times faster than the most accurate competitor. Our performance evaluation in different scenarios (environments, angles, distances) shows that Tesla generalizes well and improves the accuracy up to 20% in challenging scenarios like a through-wall setting and sensing at extreme angles. Utilizing Tesla, we develop Tesla-Rapture, a real-time implementation using a mmWave Radar on a Raspberry PI 4 and evaluate its accuracy and time-complexity. We also publish the source code, the trained models, and the implementation of the model for embedded devices.
Predicting gene expression from network topology using graph neural networks
May 08, 2020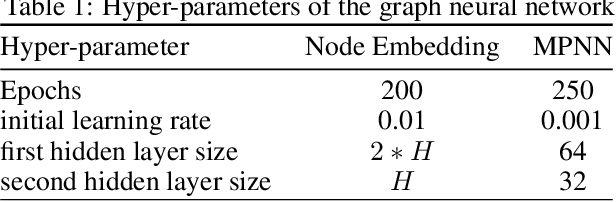
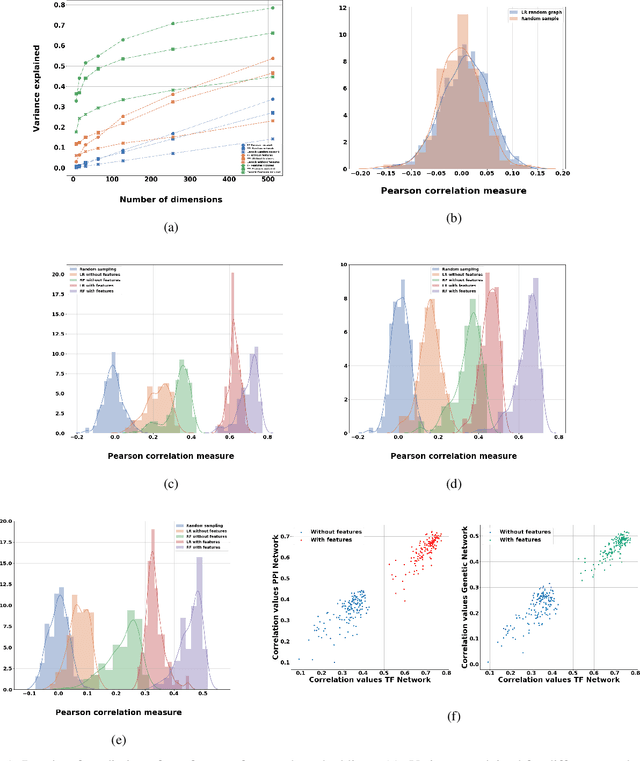
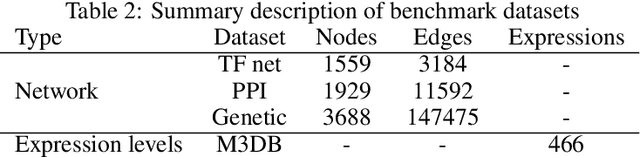
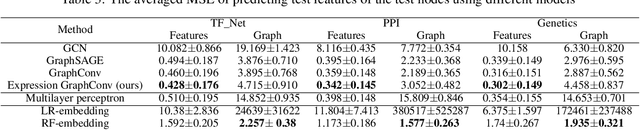
Abstract:Motivation: It is known that the structure of transcription and protein interaction networks is informative of its biological function at multiple scales. However, thus far it has not been possible to systematically connect network topology to gene expression in a quantitative way. Results: We investigated whether there is a relationship between interaction networks and gene expression values by using a graph convolutional auto-encoder and two end-to-end learning approaches for three interaction networks and hundreds of experimental conditions in the model organism \textit{E.\ coli}. Graph neural networks use a message passing framework to learn an embedding of a graph in a continuous space, either using network topology alone, or including additional node features. We found that graph embeddings trained on transcription and PPI networks can explain more than 50 and 40 percent, respectively, of the variance in gene expression data, thus confirming the relationship between network structure and gene expression value. Additionally, for the task of predicting gene expression values using GNNs, with and without additional expression training data, we found that the message passing scheme of GNNs is able to obtain the lowest mean squared error between the tested models both in prediction of unseen test values, and in an auto-encoder scheme for reconstruction of the feature matrix of expression values.
Restricted maximum-likelihood method for learning latent variance components in gene expression data with known and unknown confounders
May 06, 2020
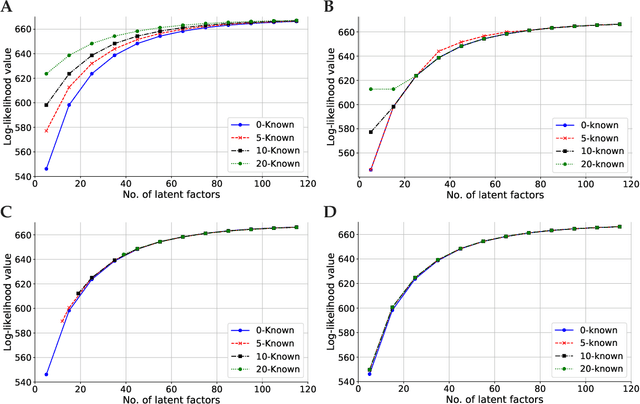


Abstract:Linear mixed modelling is a popular approach for detecting and correcting spurious sample correlations due to hidden confounders in genome-wide gene expression data. In applications where some confounding factors are known, estimating simultaneously the contribution of known and latent variance components in linear mixed models is a challenge that has so far relied on numerical gradient-based optimizers to maximize the likelihood function. This is unsatisfactory because the resulting solution is poorly characterized and the efficiency of the method may be suboptimal. Here we prove analytically that maximum-likelihood latent variables can always be chosen orthogonal to the known confounding factors, in other words, that maximum-likelihood latent variables explain sample covariances not already explained by known factors. Based on this result we propose a restricted maximum-likelihood method which estimates the latent variables by maximizing the likelihood on the restricted subspace orthogonal to the known confounding factors, and show that this reduces to probabilistic PCA on that subspace. The method then estimates the variance-covariance parameters by maximizing the remaining terms in the likelihood function given the latent variables, using a newly derived analytic solution for this problem. Compared to gradient-based optimizers, our method attains equal or higher likelihood values, can be computed using standard matrix operations, results in latent factors that don't overlap with any known factors, and has a runtime reduced by several orders of magnitude. We anticipate that the restricted maximum-likelihood method will facilitate the application of linear mixed modelling strategies for learning latent variance components to much larger gene expression datasets than currently possible.
Analytic solution and stationary phase approximation for the Bayesian lasso and elastic net
Oct 15, 2018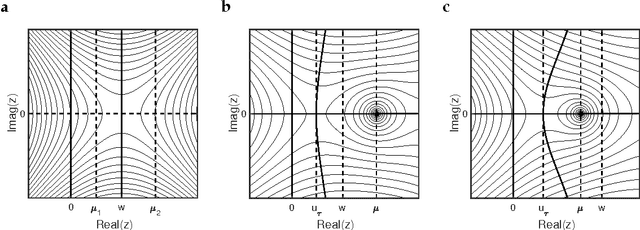

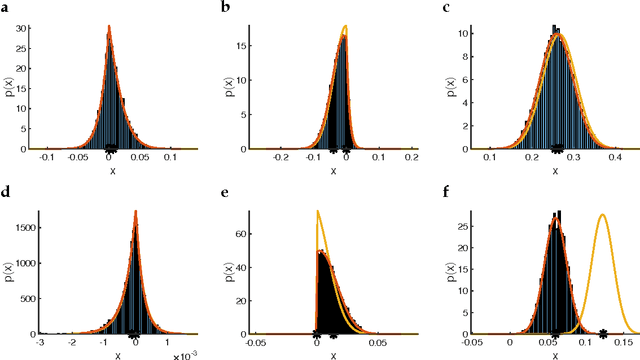
Abstract:The lasso and elastic net linear regression models impose a double-exponential prior distribution on the model parameters to achieve regression shrinkage and variable selection, allowing the inference of robust models from large data sets. However, there has been limited success in deriving estimates for the full posterior distribution of regression coefficients in these models, due to a need to evaluate analytically intractable partition function integrals. Here, the Fourier transform is used to express these integrals as complex-valued oscillatory integrals over "regression frequencies". This results in an analytic expansion and stationary phase approximation for the partition functions of the Bayesian lasso and elastic net, where the non-differentiability of the double-exponential prior has so far eluded such an approach. Use of this approximation leads to highly accurate numerical estimates for the expectation values and marginal posterior distributions of the regression coefficients, and allows for Bayesian inference of much higher dimensional models than previously possible.
Wisdom of the crowd from unsupervised dimension reduction
Nov 28, 2017



Abstract:Wisdom of the crowd, the collective intelligence derived from responses of multiple human or machine individuals to the same questions, can be more accurate than each individual, and improve social decision-making and prediction accuracy. This can also integrate multiple programs or datasets, each as an individual, for the same predictive questions. Crowd wisdom estimates each individual's independent error level arising from their limited knowledge, and finds the crowd consensus that minimizes the overall error. However, previous studies have merely built isolated, problem-specific models with limited generalizability, and mainly for binary (yes/no) responses. Here we show with simulation and real-world data that the crowd wisdom problem is analogous to one-dimensional unsupervised dimension reduction in machine learning. This provides a natural class of crowd wisdom solutions, such as principal component analysis and Isomap, which can handle binary and also continuous responses, like confidence levels, and consequently can be more accurate than existing solutions. They can even outperform supervised-learning-based collective intelligence that is calibrated on historical performance of individuals, e.g. penalized linear regression and random forest. This study unifies crowd wisdom and unsupervised dimension reduction, and thereupon introduces a broad range of highly-performing and widely-applicable crowd wisdom methods. As the costs for data acquisition and processing rapidly decrease, this study will promote and guide crowd wisdom applications in the social and natural sciences, including data fusion, meta-analysis, crowd-sourcing, and committee decision making.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge