Tim Chen
A Census-Based Genetic Algorithm for Target Set Selection Problem in Social Networks
Oct 02, 2024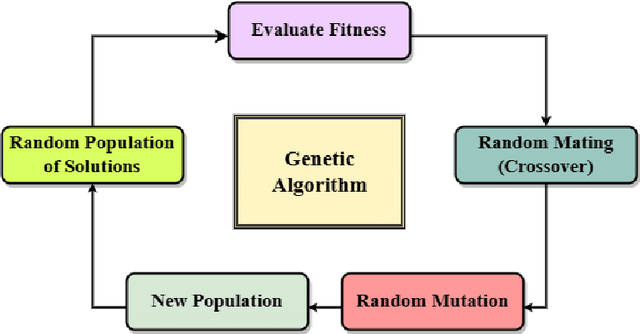
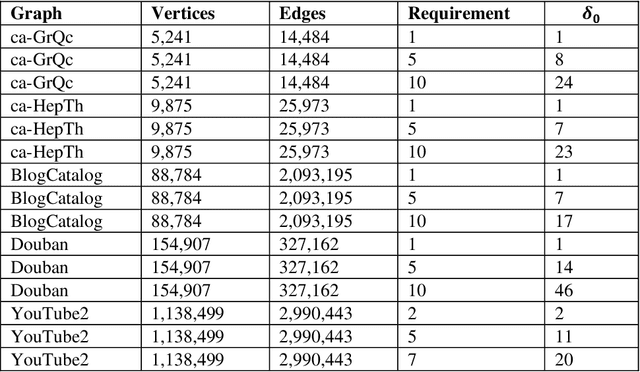
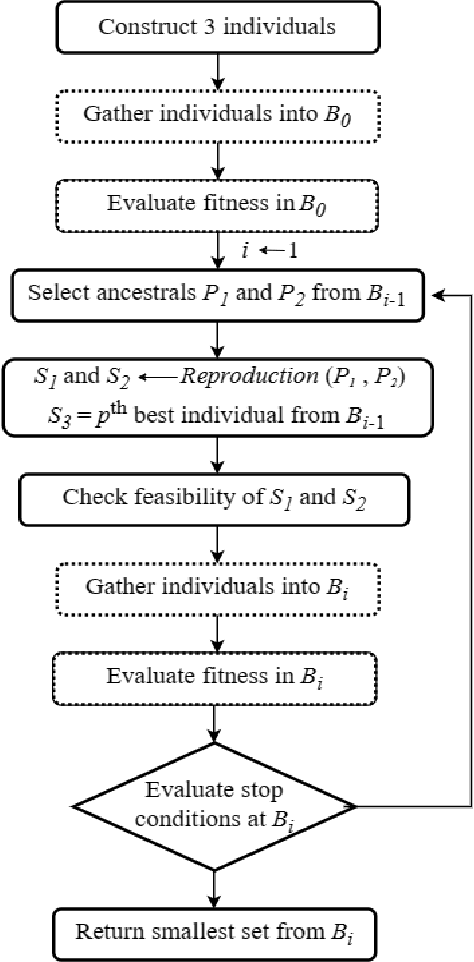
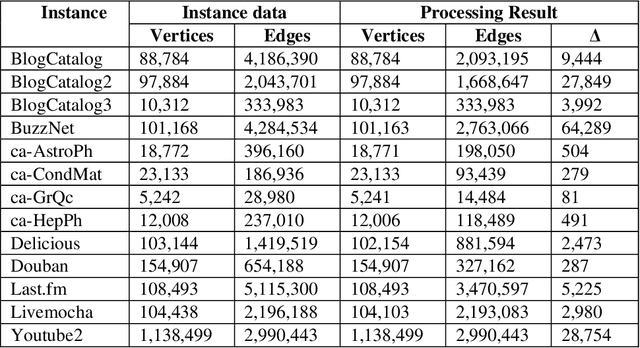
Abstract:This paper considers the Target Set Selection (TSS) Problem in social networks, a fundamental problem in viral marketing. In the TSS problem, a graph and a threshold value for each vertex of the graph are given. We need to find a minimum size vertex subset to "activate" such that all graph vertices are activated at the end of the propagation process. Specifically, we propose a novel approach called "a census-based genetic algorithm" for the TSS problem. In our algorithm, we use the idea of a census to gather and store information about each individual in a population and collect census data from the individuals constructed during the algorithm's execution so that we can achieve greater diversity and avoid premature convergence at locally optimal solutions. We use two distinct census information: (a) for each individual, the algorithm stores how many times it has been identified during the execution (b) for each network node, the algorithm counts how many times it has been included in a solution. The proposed algorithm can also self-adjust by using a parameter specifying the aggressiveness employed in each reproduction method. Additionally, the algorithm is designed to run in a parallelized environment to minimize the computational cost and check each individual's feasibility. Moreover, our algorithm finds the optimal solution in all cases while experimenting on random graphs. Furthermore, we execute the proposed algorithm on 14 large graphs of real-life social network instances from the literature, improving around 9.57 solution size (on average) and 134 vertices (in total) compared to the best solutions obtained in previous studies.
In Defense of Kalman Filtering for Polyp Tracking from Colonoscopy Videos
Jan 27, 2022
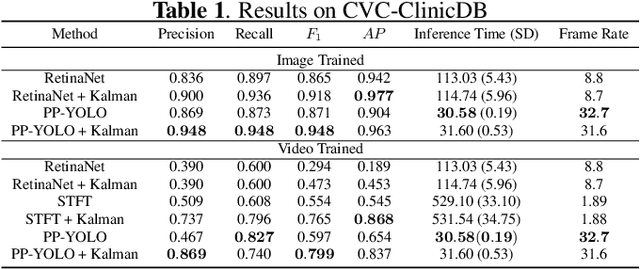
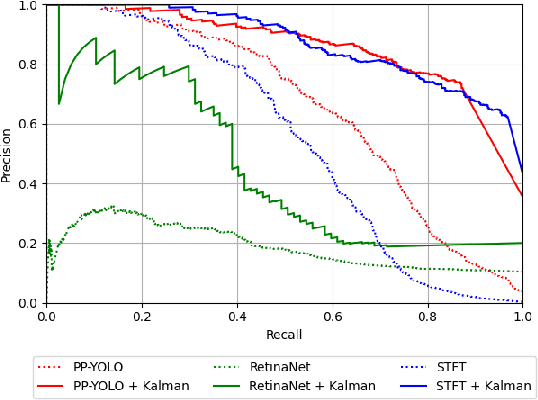
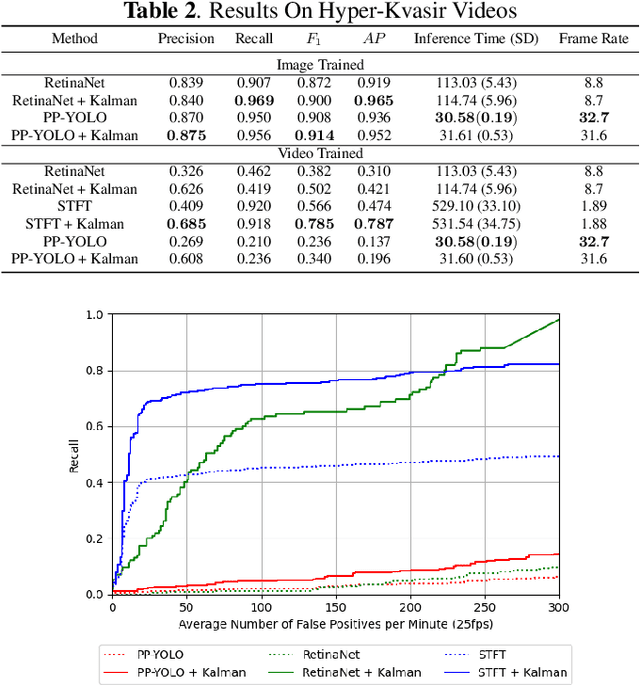
Abstract:Real-time and robust automatic detection of polyps from colonoscopy videos are essential tasks to help improve the performance of doctors during this exam. The current focus of the field is on the development of accurate but inefficient detectors that will not enable a real-time application. We advocate that the field should instead focus on the development of simple and efficient detectors that an be combined with effective trackers to allow the implementation of real-time polyp detectors. In this paper, we propose a Kalman filtering tracker that can work together with powerful, but efficient detectors, enabling the implementation of real-time polyp detectors. In particular, we show that the combination of our Kalman filtering with the detector PP-YOLO shows state-of-the-art (SOTA) detection accuracy and real-time processing. More specifically, our approach has SOTA results on the CVC-ClinicDB dataset, with a recall of 0.740, precision of 0.869, $F_1$ score of 0.799, an average precision (AP) of 0.837, and can run in real time (i.e., 30 frames per second). We also evaluate our method on a subset of the Hyper-Kvasir annotated by our clinical collaborators, resulting in SOTA results, with a recall of 0.956, precision of 0.875, $F_1$ score of 0.914, AP of 0.952, and can run in real time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge