Tiantian Yang
engGNN: A Dual-Graph Neural Network for Omics-Based Disease Classification and Feature Selection
Jan 20, 2026Abstract:Omics data, such as transcriptomics, proteomics, and metabolomics, provide critical insights into disease mechanisms and clinical outcomes. However, their high dimensionality, small sample sizes, and intricate biological networks pose major challenges for reliable prediction and meaningful interpretation. Graph Neural Networks (GNNs) offer a promising way to integrate prior knowledge by encoding feature relationships as graphs. Yet, existing methods typically rely solely on either an externally curated feature graph or a data-driven generated one, which limits their ability to capture complementary information. To address this, we propose the external and generated Graph Neural Network (engGNN), a dual-graph framework that jointly leverages both external known biological networks and data-driven generated graphs. Specifically, engGNN constructs a biologically informed undirected feature graph from established network databases and complements it with a directed feature graph derived from tree-ensemble models. This dual-graph design produces more comprehensive embeddings, thereby improving predictive performance and interpretability. Through extensive simulations and real-world applications to gene expression data, engGNN consistently outperforms state-of-the-art baselines. Beyond classification, engGNN provides interpretable feature importance scores that facilitate biologically meaningful discoveries, such as pathway enrichment analysis. Taken together, these results highlight engGNN as a robust, flexible, and interpretable framework for disease classification and biomarker discovery in high-dimensional omics contexts.
MOTGNN: Interpretable Graph Neural Networks for Multi-Omics Disease Classification
Aug 10, 2025Abstract:Integrating multi-omics data, such as DNA methylation, mRNA expression, and microRNA (miRNA) expression, offers a comprehensive view of the biological mechanisms underlying disease. However, the high dimensionality and complex interactions among omics layers present major challenges for predictive modeling. We propose Multi-Omics integration with Tree-generated Graph Neural Network (MOTGNN), a novel and interpretable framework for binary disease classification. MOTGNN employs eXtreme Gradient Boosting (XGBoost) to perform omics-specific supervised graph construction, followed by modality-specific Graph Neural Networks (GNNs) for hierarchical representation learning, and a deep feedforward network for cross-omics integration. On three real-world disease datasets, MOTGNN outperforms state-of-the-art baselines by 5-10% in accuracy, ROC-AUC, and F1-score, and remains robust to severe class imbalance (e.g., 87.2% vs. 33.4% F1 on imbalanced data). The model maintains computational efficiency through sparse graphs (2.1-2.8 edges per node) and provides built-in interpretability, revealing both top-ranked biomarkers and the relative contributions of each omics modality. These results highlight MOTGNN's potential to improve both predictive accuracy and interpretability in multi-omics disease modeling.
A Deep State Space Model for Rainfall-Runoff Simulations
Jan 24, 2025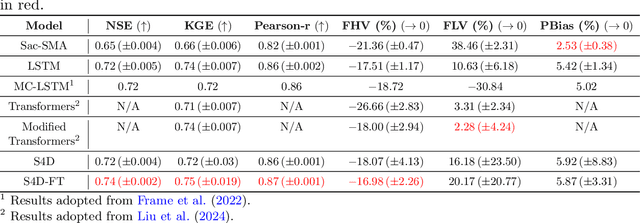

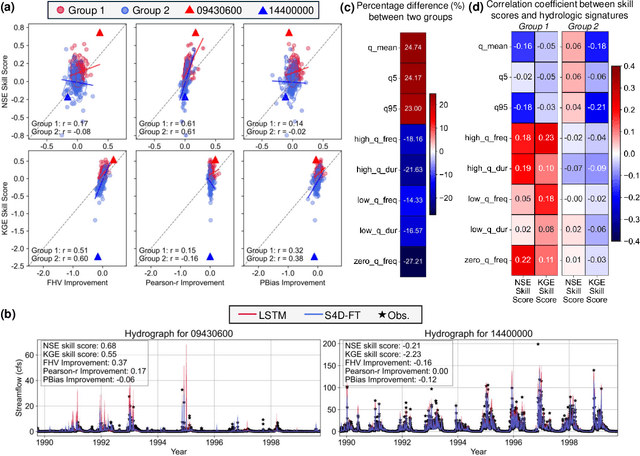
Abstract:The classical way of studying the rainfall-runoff processes in the water cycle relies on conceptual or physically-based hydrologic models. Deep learning (DL) has recently emerged as an alternative and blossomed in hydrology community for rainfall-runoff simulations. However, the decades-old Long Short-Term Memory (LSTM) network remains the benchmark for this task, outperforming newer architectures like Transformers. In this work, we propose a State Space Model (SSM), specifically the Frequency Tuned Diagonal State Space Sequence (S4D-FT) model, for rainfall-runoff simulations. The proposed S4D-FT is benchmarked against the established LSTM and a physically-based Sacramento Soil Moisture Accounting model across 531 watersheds in the contiguous United States (CONUS). Results show that S4D-FT is able to outperform the LSTM model across diverse regions. Our pioneering introduction of the S4D-FT for rainfall-runoff simulations challenges the dominance of LSTM in the hydrology community and expands the arsenal of DL tools available for hydrological modeling.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge