Taojun Hu
Unveiling LLM Evaluation Focused on Metrics: Challenges and Solutions
Apr 14, 2024Abstract:Natural Language Processing (NLP) is witnessing a remarkable breakthrough driven by the success of Large Language Models (LLMs). LLMs have gained significant attention across academia and industry for their versatile applications in text generation, question answering, and text summarization. As the landscape of NLP evolves with an increasing number of domain-specific LLMs employing diverse techniques and trained on various corpus, evaluating performance of these models becomes paramount. To quantify the performance, it's crucial to have a comprehensive grasp of existing metrics. Among the evaluation, metrics which quantifying the performance of LLMs play a pivotal role. This paper offers a comprehensive exploration of LLM evaluation from a metrics perspective, providing insights into the selection and interpretation of metrics currently in use. Our main goal is to elucidate their mathematical formulations and statistical interpretations. We shed light on the application of these metrics using recent Biomedical LLMs. Additionally, we offer a succinct comparison of these metrics, aiding researchers in selecting appropriate metrics for diverse tasks. The overarching goal is to furnish researchers with a pragmatic guide for effective LLM evaluation and metric selection, thereby advancing the understanding and application of these large language models.
ADRNet: A Generalized Collaborative Filtering Framework Combining Clinical and Non-Clinical Data for Adverse Drug Reaction Prediction
Aug 03, 2023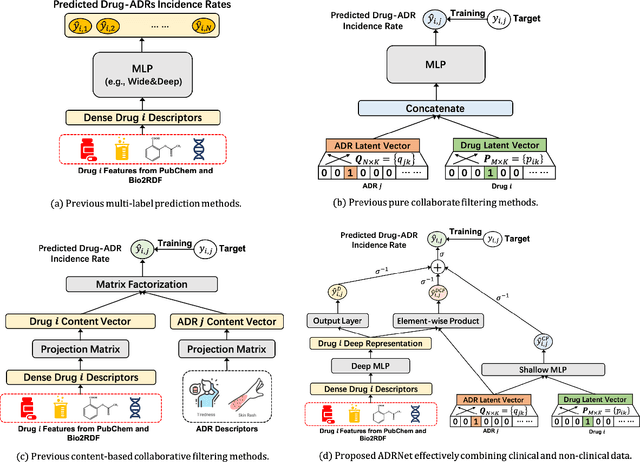
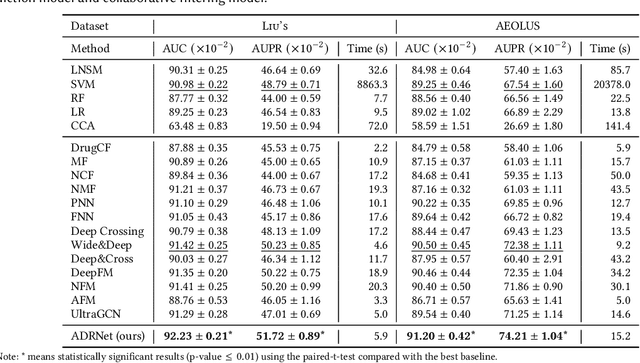
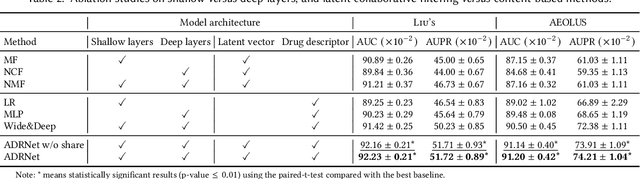
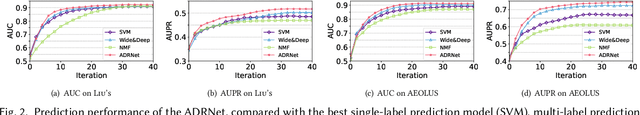
Abstract:Adverse drug reaction (ADR) prediction plays a crucial role in both health care and drug discovery for reducing patient mortality and enhancing drug safety. Recently, many studies have been devoted to effectively predict the drug-ADRs incidence rates. However, these methods either did not effectively utilize non-clinical data, i.e., physical, chemical, and biological information about the drug, or did little to establish a link between content-based and pure collaborative filtering during the training phase. In this paper, we first formulate the prediction of multi-label ADRs as a drug-ADR collaborative filtering problem, and to the best of our knowledge, this is the first work to provide extensive benchmark results of previous collaborative filtering methods on two large publicly available clinical datasets. Then, by exploiting the easy accessible drug characteristics from non-clinical data, we propose ADRNet, a generalized collaborative filtering framework combining clinical and non-clinical data for drug-ADR prediction. Specifically, ADRNet has a shallow collaborative filtering module and a deep drug representation module, which can exploit the high-dimensional drug descriptors to further guide the learning of low-dimensional ADR latent embeddings, which incorporates both the benefits of collaborative filtering and representation learning. Extensive experiments are conducted on two publicly available real-world drug-ADR clinical datasets and two non-clinical datasets to demonstrate the accuracy and efficiency of the proposed ADRNet. The code is available at https://github.com/haoxuanli-pku/ADRnet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge