Suthesh Sivapalaratnam
Deep Generative Classification of Blood Cell Morphology
Aug 16, 2024



Abstract:Accurate classification of haematological cells is critical for diagnosing blood disorders, but presents significant challenges for machine automation owing to the complexity of cell morphology, heterogeneities of biological, pathological, and imaging characteristics, and the imbalance of cell type frequencies. We introduce CytoDiffusion, a diffusion-based classifier that effectively models blood cell morphology, combining accurate classification with robust anomaly detection, resistance to distributional shifts, interpretability, data efficiency, and superhuman uncertainty quantification. Our approach outperforms state-of-the-art discriminative models in anomaly detection (AUC 0.976 vs. 0.919), resistance to domain shifts (85.85% vs. 74.38% balanced accuracy), and performance in low-data regimes (95.88% vs. 94.95% balanced accuracy). Notably, our model generates synthetic blood cell images that are nearly indistinguishable from real images, as demonstrated by a Turing test in which expert haematologists achieved only 52.3% accuracy (95% CI: [50.5%, 54.2%]). Furthermore, we enhance model explainability through the generation of directly interpretable counterfactual heatmaps. Our comprehensive evaluation framework, encompassing these multiple performance dimensions, establishes a new benchmark for medical image analysis in haematology, ultimately enabling improved diagnostic accuracy in clinical settings. Our code is available at https://github.com/Deltadahl/CytoDiffusion.
Dis-AE: Multi-domain & Multi-task Generalisation on Real-World Clinical Data
Jun 15, 2023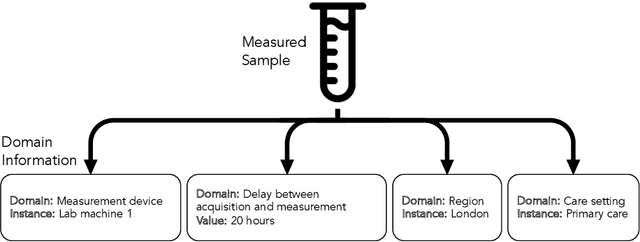
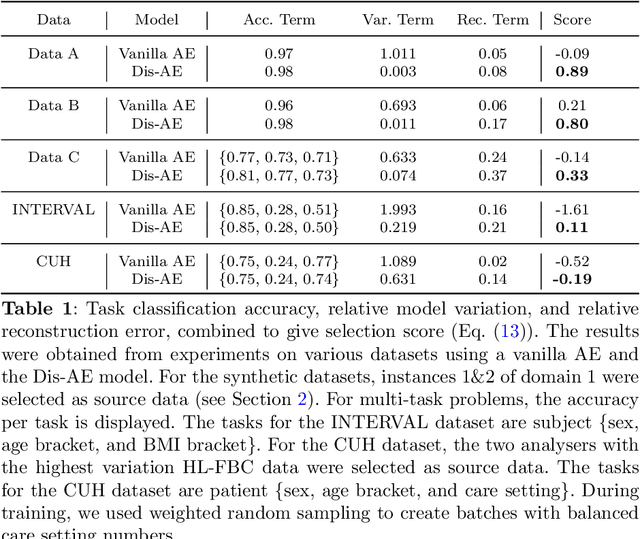
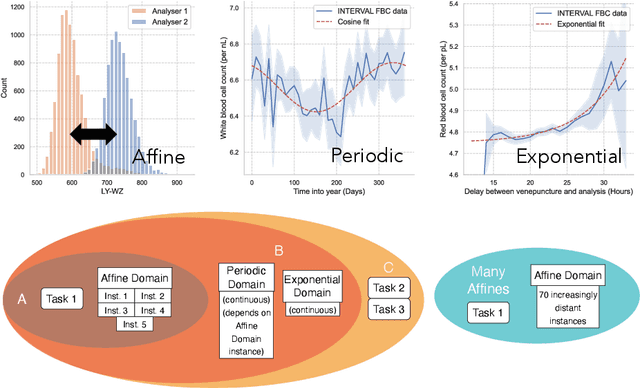
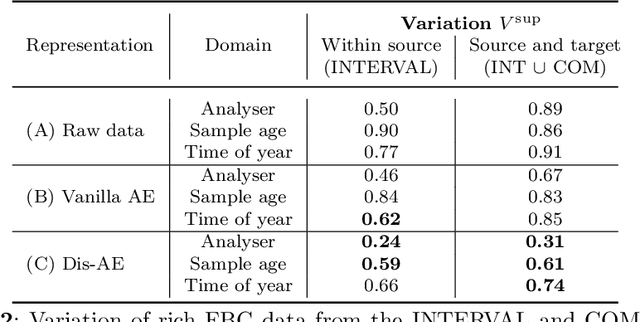
Abstract:Clinical data is often affected by clinically irrelevant factors such as discrepancies between measurement devices or differing processing methods between sites. In the field of machine learning (ML), these factors are known as domains and the distribution differences they cause in the data are known as domain shifts. ML models trained using data from one domain often perform poorly when applied to data from another domain, potentially leading to wrong predictions. As such, developing machine learning models that can generalise well across multiple domains is a challenging yet essential task in the successful application of ML in clinical practice. In this paper, we propose a novel disentangled autoencoder (Dis-AE) neural network architecture that can learn domain-invariant data representations for multi-label classification of medical measurements even when the data is influenced by multiple interacting domain shifts at once. The model utilises adversarial training to produce data representations from which the domain can no longer be determined. We evaluate the model's domain generalisation capabilities on synthetic datasets and full blood count (FBC) data from blood donors as well as primary and secondary care patients, showing that Dis-AE improves model generalisation on multiple domains simultaneously while preserving clinically relevant information.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge