Sundaresh Ram
CellGenNet: A Knowledge-Distilled Framework for Robust Cell Segmentation in Cancer Tissues
Nov 19, 2025Abstract:Accurate nuclei segmentation in microscopy whole slide images (WSIs) remains challenging due to variability in staining, imaging conditions, and tissue morphology. We propose CellGenNet, a knowledge distillation framework for robust cross-tissue cell segmentation under limited supervision. CellGenNet adopts a student-teacher architecture, where a capacity teacher is trained on sparse annotations and generates soft pseudo-labels for unlabeled regions. The student is optimized using a joint objective that integrates ground-truth labels, teacher-derived probabilistic targets, and a hybrid loss function combining binary cross-entropy and Tversky loss, enabling asymmetric penalties to mitigate class imbalance and better preserve minority nuclear structures. Consistency regularization and layerwise dropout further stabilize feature representations and promote reliable feature transfer. Experiments across diverse cancer tissue WSIs show that CellGenNet improves segmentation accuracy and generalization over supervised and semi-supervised baselines, supporting scalable and reproducible histopathology analysis.
RepAir: A Framework for Airway Segmentation and Discontinuity Correction in CT
Nov 18, 2025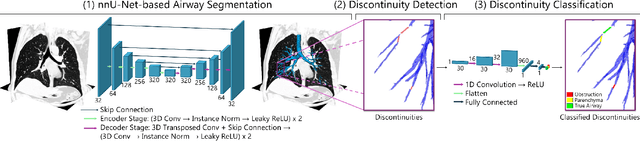
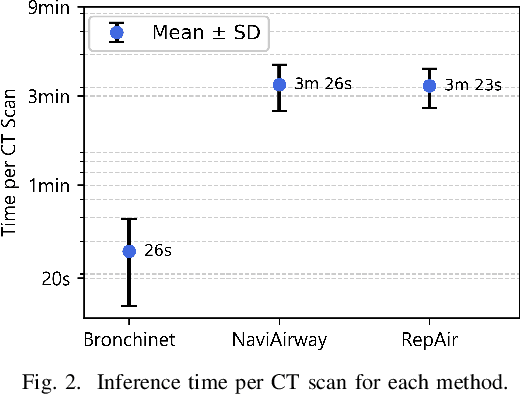
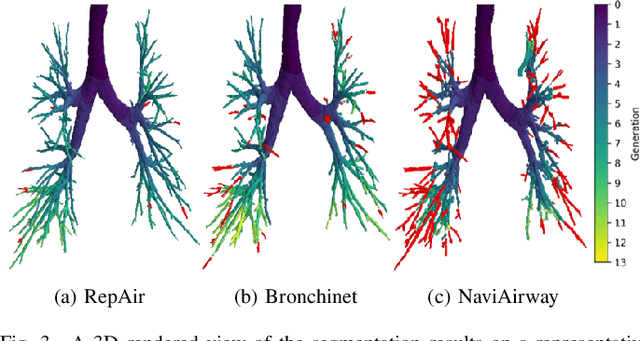
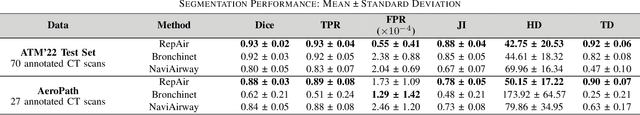
Abstract:Accurate airway segmentation from chest computed tomography (CT) scans is essential for quantitative lung analysis, yet manual annotation is impractical and many automated U-Net-based methods yield disconnected components that hinder reliable biomarker extraction. We present RepAir, a three-stage framework for robust 3D airway segmentation that combines an nnU-Net-based network with anatomically informed topology correction. The segmentation network produces an initial airway mask, after which a skeleton-based algorithm identifies potential discontinuities and proposes reconnections. A 1D convolutional classifier then determines which candidate links correspond to true anatomical branches versus false or obstructed paths. We evaluate RepAir on two distinct datasets: ATM'22, comprising annotated CT scans from predominantly healthy subjects and AeroPath, encompassing annotated scans with severe airway pathology. Across both datasets, RepAir outperforms existing 3D U-Net-based approaches such as Bronchinet and NaviAirway on both voxel-level and topological metrics, and produces more complete and anatomically consistent airway trees while maintaining high segmentation accuracy.
Lung Cancer Lesion Detection in Histopathology Images Using Graph-Based Sparse PCA Network
Oct 27, 2021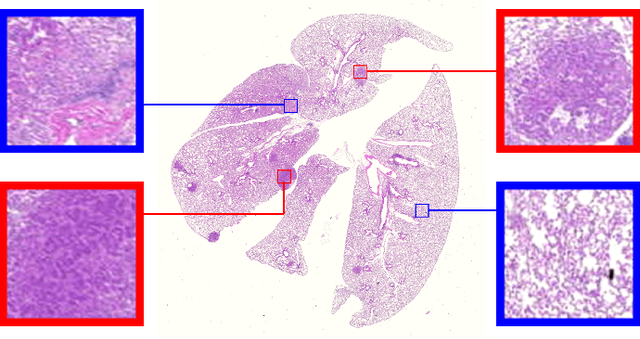
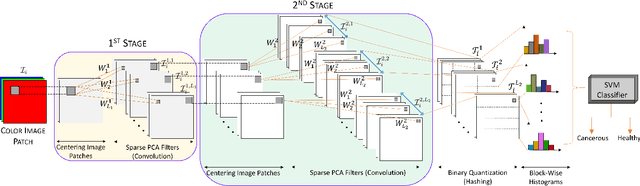
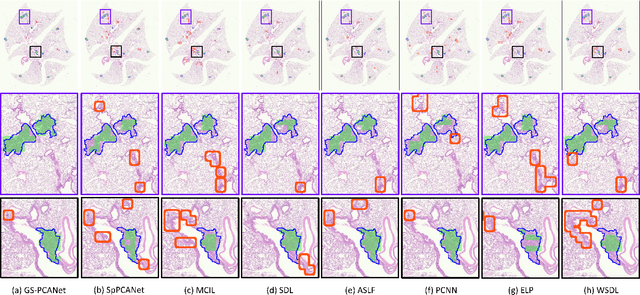
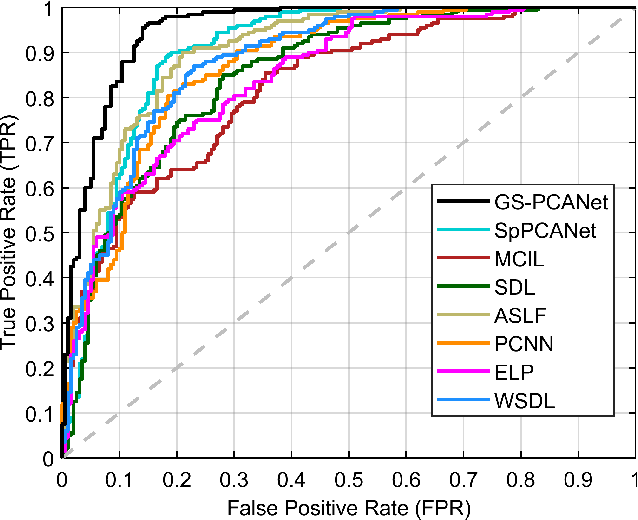
Abstract:Early detection of lung cancer is critical for improvement of patient survival. To address the clinical need for efficacious treatments, genetically engineered mouse models (GEMM) have become integral in identifying and evaluating the molecular underpinnings of this complex disease that may be exploited as therapeutic targets. Assessment of GEMM tumor burden on histopathological sections performed by manual inspection is both time consuming and prone to subjective bias. Therefore, an interplay of needs and challenges exists for computer-aided diagnostic tools, for accurate and efficient analysis of these histopathology images. In this paper, we propose a simple machine learning approach called the graph-based sparse principal component analysis (GS-PCA) network, for automated detection of cancerous lesions on histological lung slides stained by hematoxylin and eosin (H&E). Our method comprises four steps: 1) cascaded graph-based sparse PCA, 2) PCA binary hashing, 3) block-wise histograms, and 4) support vector machine (SVM) classification. In our proposed architecture, graph-based sparse PCA is employed to learn the filter banks of the multiple stages of a convolutional network. This is followed by PCA hashing and block histograms for indexing and pooling. The meaningful features extracted from this GS-PCA are then fed to an SVM classifier. We evaluate the performance of the proposed algorithm on H&E slides obtained from an inducible K-rasG12D lung cancer mouse model using precision/recall rates, F-score, Tanimoto coefficient, and area under the curve (AUC) of the receiver operator characteristic (ROC) and show that our algorithm is efficient and provides improved detection accuracy compared to existing algorithms.
Robust Segmentation of Cell Nuclei in 3-D Microscopy Images
Oct 07, 2021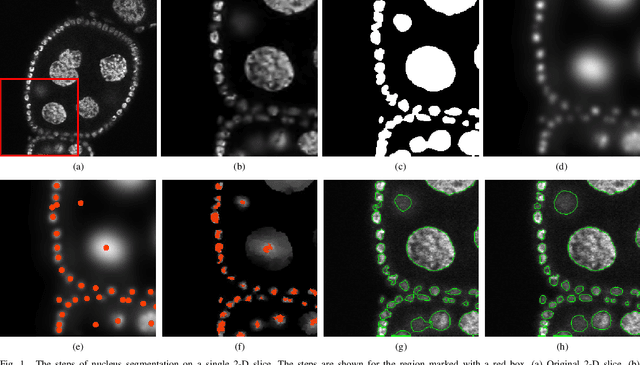
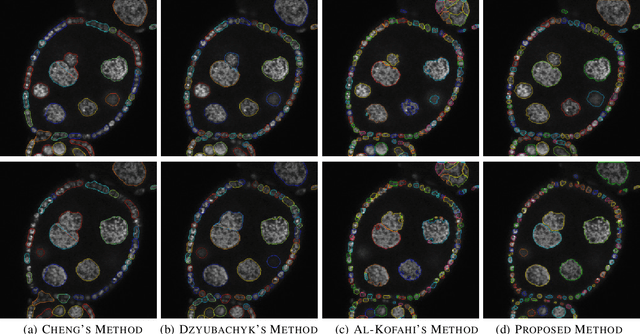
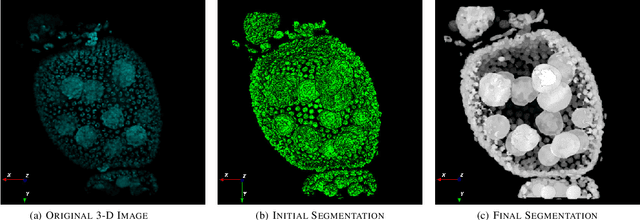
Abstract:Accurate segmentation of 3-D cell nuclei in microscopy images is essential for the study of nuclear organization, gene expression, and cell morphodynamics. Current image segmentation methods are challenged by the complexity and variability of microscopy images and often over-segment or under-segment the cell nuclei. Thus, there is a need to improve segmentation accuracy and reliability, as well as the level of automation. In this paper, we propose a new automated algorithm for robust segmentation of 3-D cell nuclei using the concepts of random walk, graph theory, and mathematical morphology as the foundation. Like other segmentation algorithms, we first use a seed detection/marker extraction algorithm to find a seed voxel for each individual cell nucleus. Next, using the concept of random walk on a graph we find the probability of all the pixels in the 3-D image to reach the seed pixels of each nucleus identified by the seed detection algorithm. We then generate a 3-D response image by combining these probabilities for each voxel and use the marker controlled watershed transform on this response image to obtain an initial segmentation of the cell nuclei. Finally, we apply local region-based active contours to obtain final segmentation of the cell nuclei. The advantage of using such an approach is that it is capable of accurately segmenting highly textured cells having inhomogeneous intensities and varying shapes and sizes. The proposed algorithm was compared with three other automated nucleus segmentation algorithms for segmentation accuracy using overlap measure, Tanimoto index, Rand index, F-score, and Hausdorff distance measure. Quantitative and qualitative results show that our algorithm provides improved segmentation accuracy compared to existing algorithms.
Object sieving and morphological closing to reduce false detections in wide-area aerial imagery
Oct 28, 2020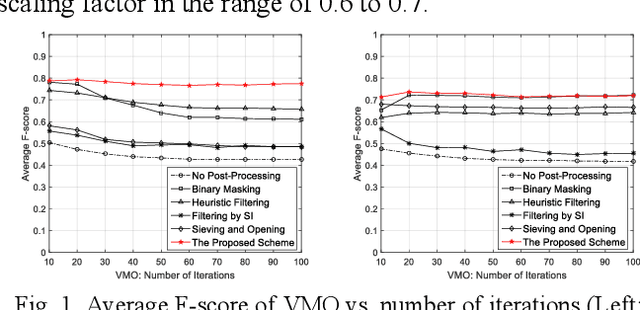
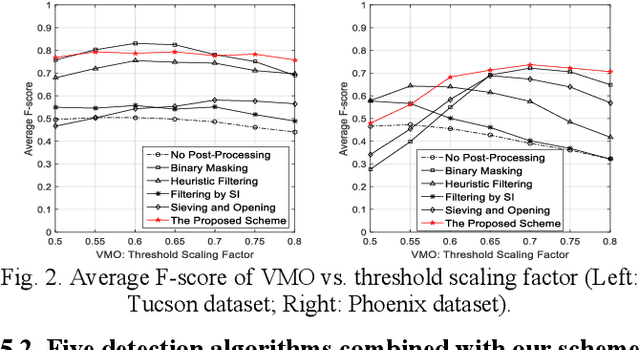
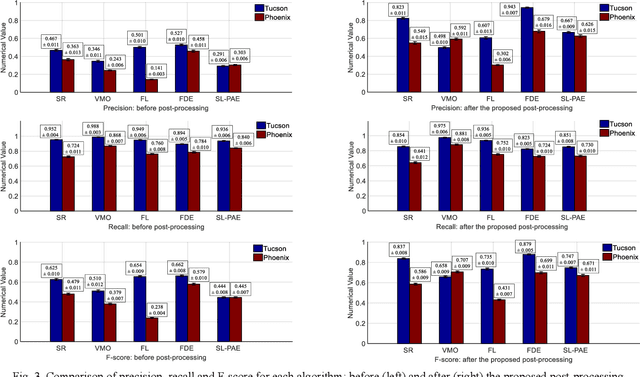
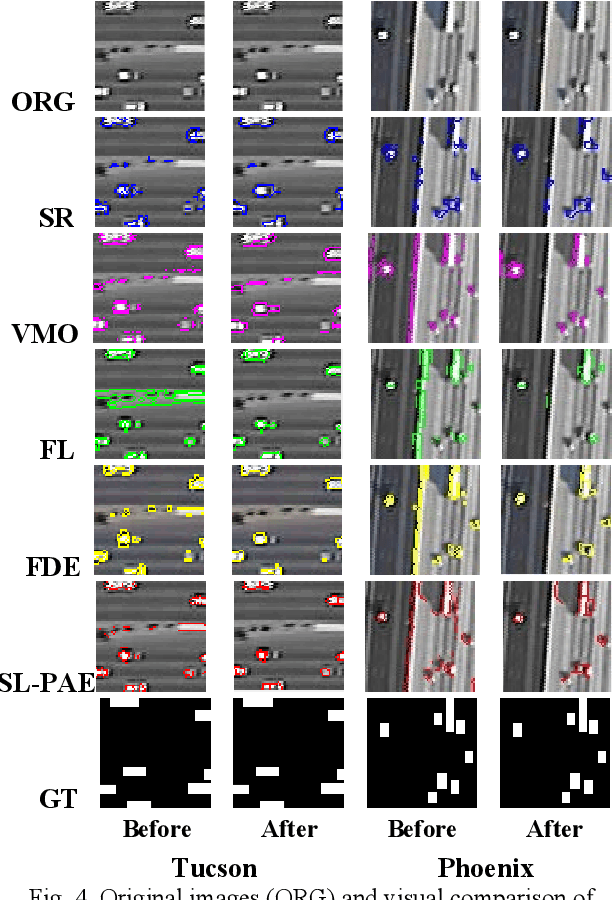
Abstract:For object detection in wide-area aerial imagery, post-processing is usually needed to reduce false detections. We propose a two-stage post-processing scheme which comprises an area-thresholding sieving process and a morphological closing operation. We use two wide-area aerial videos to compare the performance of five object detection algorithms in the absence and in the presence of our post-processing scheme. The automatic detection results are compared with the ground-truth objects. Several metrics are used for performance comparison.
Drive-Net: Convolutional Network for Driver Distraction Detection
Jun 22, 2020
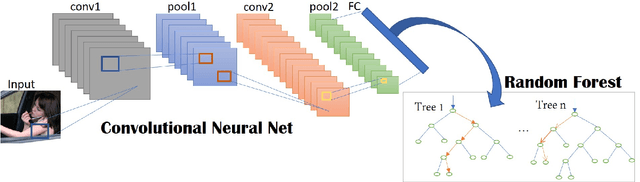
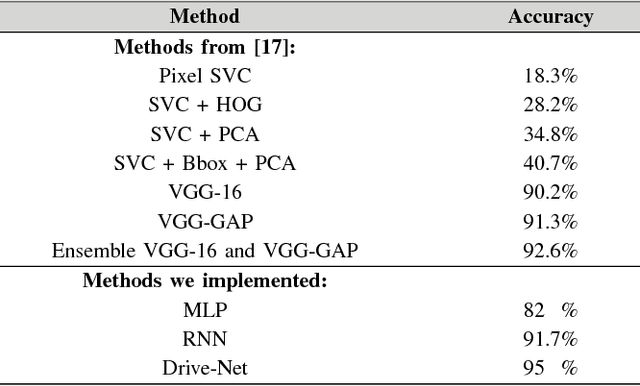
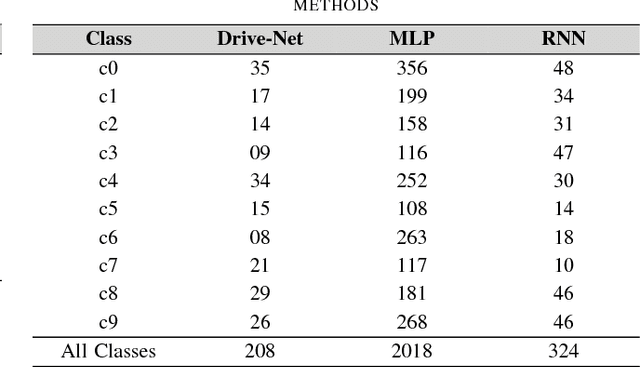
Abstract:To help prevent motor vehicle accidents, there has been significant interest in finding an automated method to recognize signs of driver distraction, such as talking to passengers, fixing hair and makeup, eating and drinking, and using a mobile phone. In this paper, we present an automated supervised learning method called Drive-Net for driver distraction detection. Drive-Net uses a combination of a convolutional neural network (CNN) and a random decision forest for classifying images of a driver. We compare the performance of our proposed Drive-Net to two other popular machine-learning approaches: a recurrent neural network (RNN), and a multi-layer perceptron (MLP). We test the methods on a publicly available database of images acquired under a controlled environment containing about 22425 images manually annotated by an expert. Results show that Drive-Net achieves a detection accuracy of 95%, which is 2% more than the best results obtained on the same database using other methods
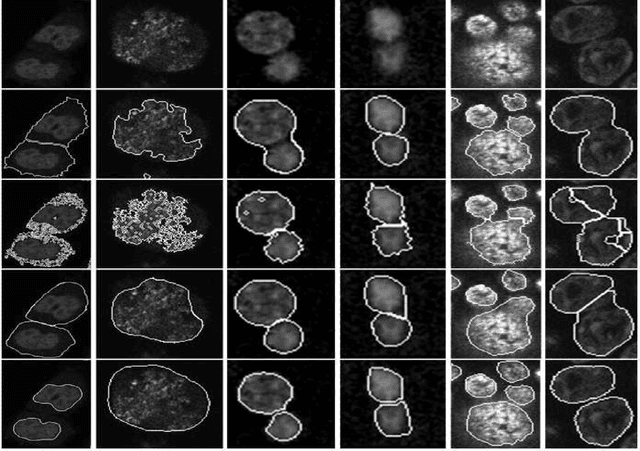
Joint Cell Nuclei Detection and Segmentation in Microscopy Images Using 3D Convolutional Networks
Sep 06, 2018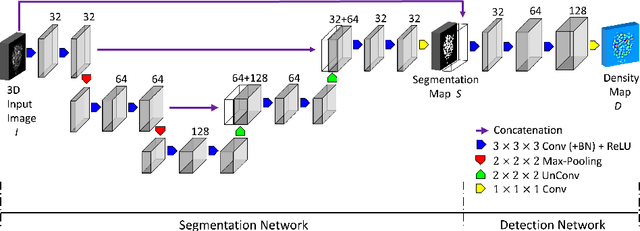
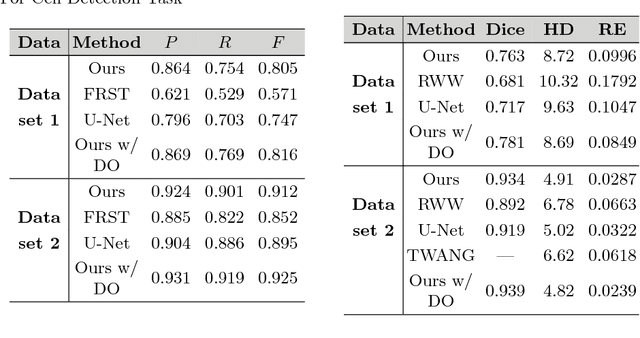
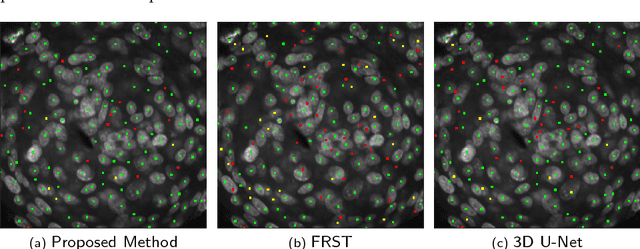
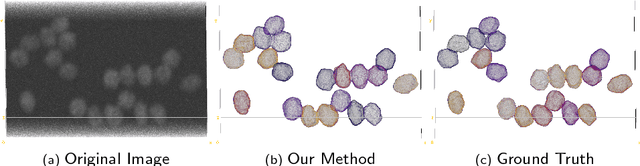
Abstract:We propose a 3D convolutional neural network to simultaneously segment and detect cell nuclei in confocal microscopy images. Mirroring the co-dependency of these tasks, our proposed model consists of two serial components: the first part computes a segmentation of cell bodies, while the second module identifies the centers of these cells. Our model is trained end-to-end from scratch on a mouse parotid salivary gland stem cell nuclei dataset comprising 107 image stacks from three independent cell preparations, each containing several hundred individual cell nuclei in 3D. In our experiments, we conduct a thorough evaluation of both detection accuracy and segmentation quality, on two different datasets. The results show that the proposed method provides significantly improved detection and segmentation accuracy compared to state-of-the-art and benchmark algorithms. Finally, we use a previously described test-time drop-out strategy to obtain uncertainty estimates on our predictions and validate these estimates by demonstrating that they are strongly correlated with accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge