Jeffrey J. Rodriguez
Crowd-Certain: Label Aggregation in Crowdsourced and Ensemble Learning Classification
Oct 25, 2023Abstract:Crowdsourcing systems have been used to accumulate massive amounts of labeled data for applications such as computer vision and natural language processing. However, because crowdsourced labeling is inherently dynamic and uncertain, developing a technique that can work in most situations is extremely challenging. In this paper, we introduce Crowd-Certain, a novel approach for label aggregation in crowdsourced and ensemble learning classification tasks that offers improved performance and computational efficiency for different numbers of annotators and a variety of datasets. The proposed method uses the consistency of the annotators versus a trained classifier to determine a reliability score for each annotator. Furthermore, Crowd-Certain leverages predicted probabilities, enabling the reuse of trained classifiers on future sample data, thereby eliminating the need for recurrent simulation processes inherent in existing methods. We extensively evaluated our approach against ten existing techniques across ten different datasets, each labeled by varying numbers of annotators. The findings demonstrate that Crowd-Certain outperforms the existing methods (Tao, Sheng, KOS, MACE, MajorityVote, MMSR, Wawa, Zero-Based Skill, GLAD, and Dawid Skene), in nearly all scenarios, delivering higher average accuracy, F1 scores, and AUC rates. Additionally, we introduce a variation of two existing confidence score measurement techniques. Finally we evaluate these two confidence score techniques using two evaluation metrics: Expected Calibration Error (ECE) and Brier Score Loss. Our results show that Crowd-Certain achieves higher Brier Score, and lower ECE across the majority of the examined datasets, suggesting better calibrated results.
Lung Cancer Lesion Detection in Histopathology Images Using Graph-Based Sparse PCA Network
Oct 27, 2021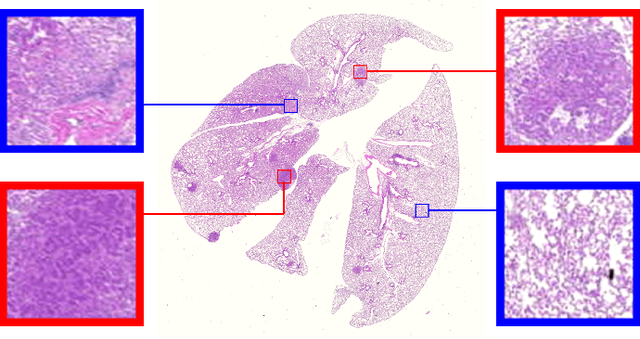
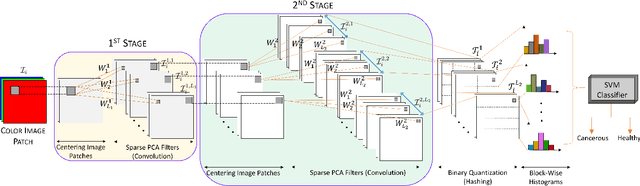
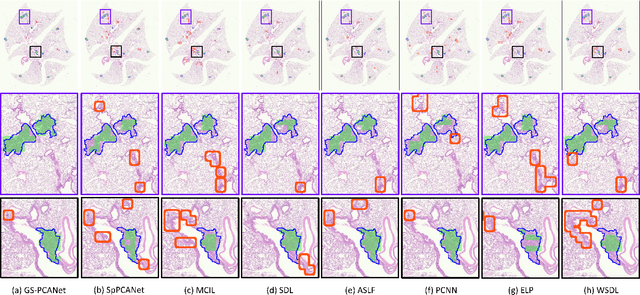
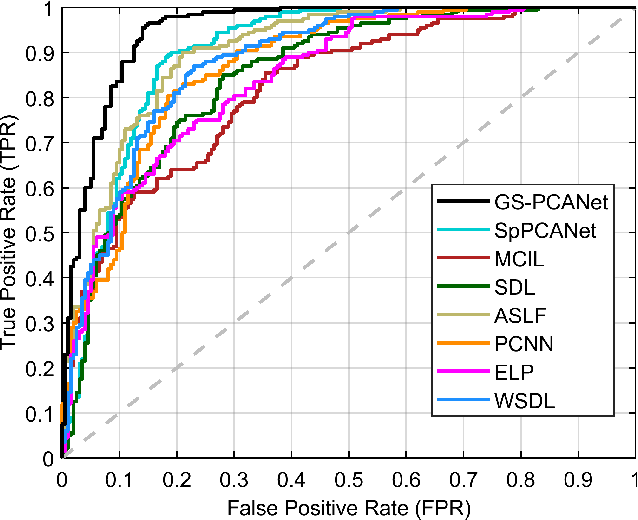
Abstract:Early detection of lung cancer is critical for improvement of patient survival. To address the clinical need for efficacious treatments, genetically engineered mouse models (GEMM) have become integral in identifying and evaluating the molecular underpinnings of this complex disease that may be exploited as therapeutic targets. Assessment of GEMM tumor burden on histopathological sections performed by manual inspection is both time consuming and prone to subjective bias. Therefore, an interplay of needs and challenges exists for computer-aided diagnostic tools, for accurate and efficient analysis of these histopathology images. In this paper, we propose a simple machine learning approach called the graph-based sparse principal component analysis (GS-PCA) network, for automated detection of cancerous lesions on histological lung slides stained by hematoxylin and eosin (H&E). Our method comprises four steps: 1) cascaded graph-based sparse PCA, 2) PCA binary hashing, 3) block-wise histograms, and 4) support vector machine (SVM) classification. In our proposed architecture, graph-based sparse PCA is employed to learn the filter banks of the multiple stages of a convolutional network. This is followed by PCA hashing and block histograms for indexing and pooling. The meaningful features extracted from this GS-PCA are then fed to an SVM classifier. We evaluate the performance of the proposed algorithm on H&E slides obtained from an inducible K-rasG12D lung cancer mouse model using precision/recall rates, F-score, Tanimoto coefficient, and area under the curve (AUC) of the receiver operator characteristic (ROC) and show that our algorithm is efficient and provides improved detection accuracy compared to existing algorithms.
Robust Segmentation of Cell Nuclei in 3-D Microscopy Images
Oct 07, 2021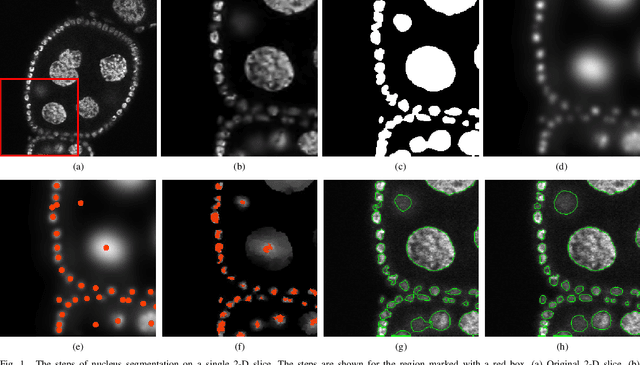
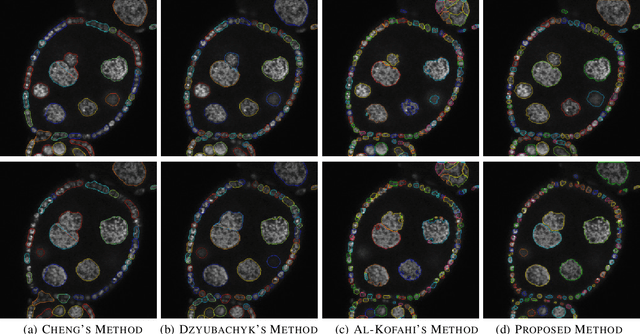
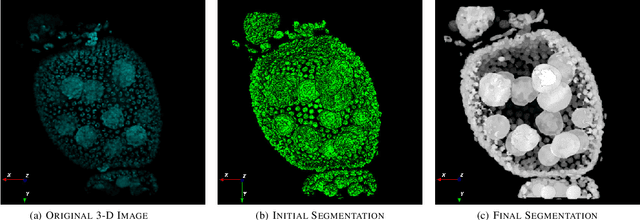
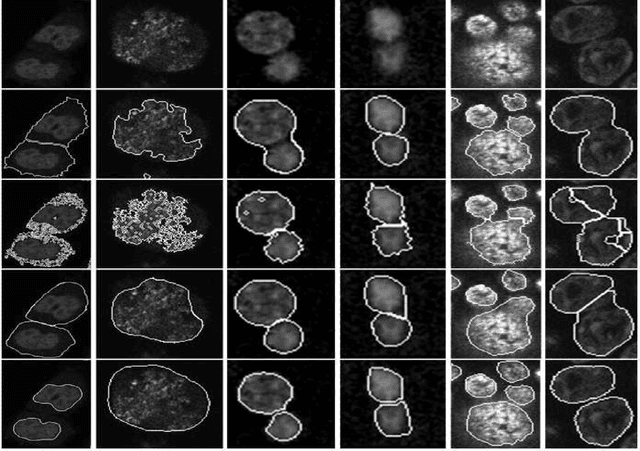
Abstract:Accurate segmentation of 3-D cell nuclei in microscopy images is essential for the study of nuclear organization, gene expression, and cell morphodynamics. Current image segmentation methods are challenged by the complexity and variability of microscopy images and often over-segment or under-segment the cell nuclei. Thus, there is a need to improve segmentation accuracy and reliability, as well as the level of automation. In this paper, we propose a new automated algorithm for robust segmentation of 3-D cell nuclei using the concepts of random walk, graph theory, and mathematical morphology as the foundation. Like other segmentation algorithms, we first use a seed detection/marker extraction algorithm to find a seed voxel for each individual cell nucleus. Next, using the concept of random walk on a graph we find the probability of all the pixels in the 3-D image to reach the seed pixels of each nucleus identified by the seed detection algorithm. We then generate a 3-D response image by combining these probabilities for each voxel and use the marker controlled watershed transform on this response image to obtain an initial segmentation of the cell nuclei. Finally, we apply local region-based active contours to obtain final segmentation of the cell nuclei. The advantage of using such an approach is that it is capable of accurately segmenting highly textured cells having inhomogeneous intensities and varying shapes and sizes. The proposed algorithm was compared with three other automated nucleus segmentation algorithms for segmentation accuracy using overlap measure, Tanimoto index, Rand index, F-score, and Hausdorff distance measure. Quantitative and qualitative results show that our algorithm provides improved segmentation accuracy compared to existing algorithms.
Object sieving and morphological closing to reduce false detections in wide-area aerial imagery
Oct 28, 2020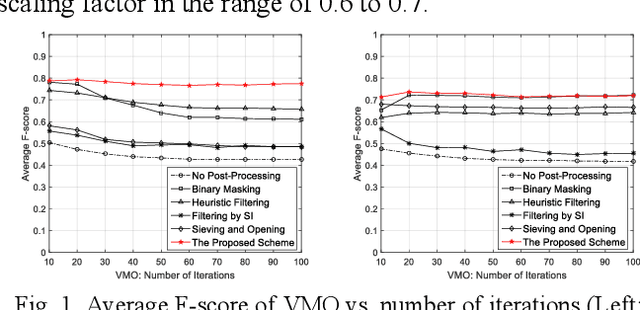
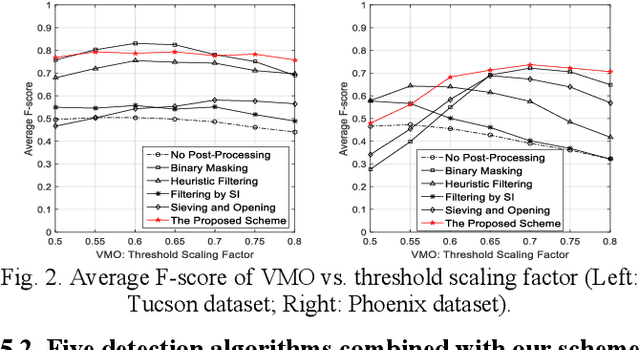
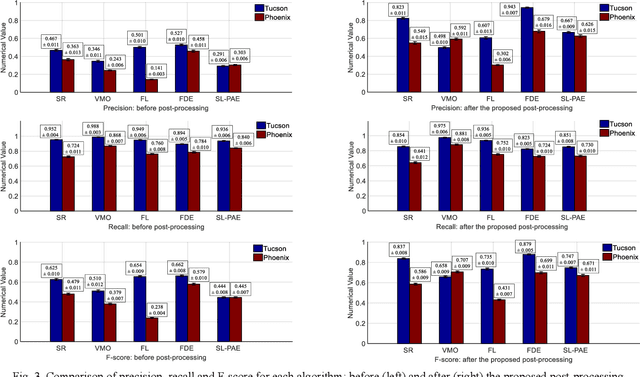
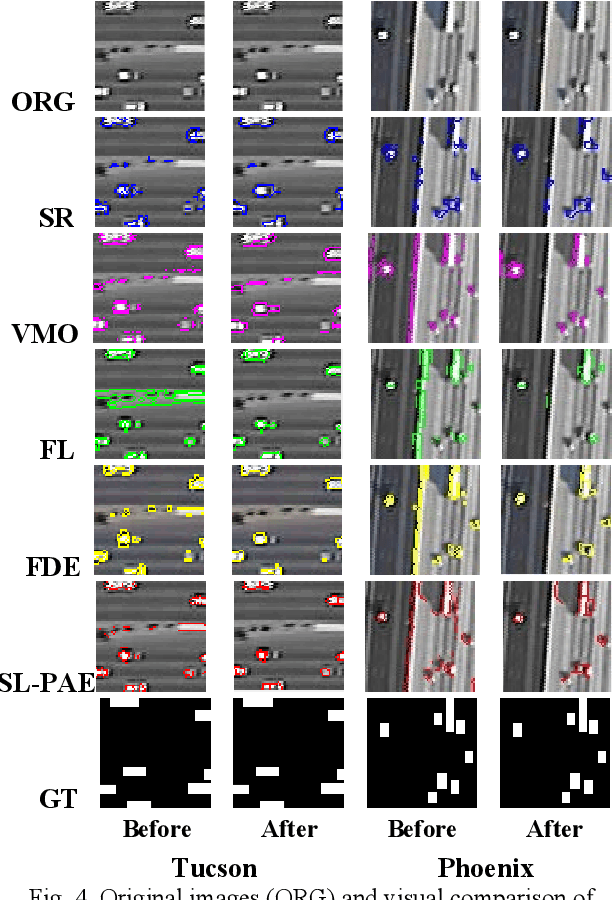
Abstract:For object detection in wide-area aerial imagery, post-processing is usually needed to reduce false detections. We propose a two-stage post-processing scheme which comprises an area-thresholding sieving process and a morphological closing operation. We use two wide-area aerial videos to compare the performance of five object detection algorithms in the absence and in the presence of our post-processing scheme. The automatic detection results are compared with the ground-truth objects. Several metrics are used for performance comparison.
Deep learning classification of chest x-ray images
Jun 22, 2020



Abstract:We propose a deep learning based method for classification of commonly occurring pathologies in chest X-ray images. The vast number of publicly available chest X-ray images provides the data necessary for successfully employing deep learning methodologies to reduce the misdiagnosis of thoracic diseases. We applied our method to the classification of two example pathologies, pulmonary nodules and cardiomegaly, and we compared the performance of our method to three existing methods. The results show an improvement in AUC for detection of nodules and cardiomegaly compared to the existing methods.
* 4 pages, 4 figures, 2 tables, conference , SSIAI 2020
A Conditional Random Field Model for Context Aware Cloud Detection in Sky Images
Jun 18, 2019



Abstract:A conditional random field (CRF) model for cloud detection in ground based sky images is presented. We show that very high cloud detection accuracy can be achieved by combining a discriminative classifier and a higher order clique potential in a CRF framework. The image is first divided into homogeneous regions using a mean shift clustering algorithm and then a CRF model is defined over these regions. The various parameters involved are estimated using training data and the inference is performed using Iterated Conditional Modes (ICM) algorithm. We demonstrate how taking spatial context into account can boost the accuracy. We present qualitative and quantitative results to prove the superior performance of this framework in comparison with other state of the art methods applied for cloud detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge