Soumyaroop Nandi
BioTamperNet: Affinity-Guided State-Space Model Detecting Tampered Biomedical Images
Feb 01, 2026Abstract:We propose BioTamperNet, a novel framework for detecting duplicated regions in tampered biomedical images, leveraging affinity-guided attention inspired by State Space Model (SSM) approximations. Existing forensic models, primarily trained on natural images, often underperform on biomedical data where subtle manipulations can compromise experimental validity. To address this, BioTamperNet introduces an affinity-guided self-attention module to capture intra-image similarities and an affinity-guided cross-attention module to model cross-image correspondences. Our design integrates lightweight SSM-inspired linear attention mechanisms to enable efficient, fine-grained localization. Trained end-to-end, BioTamperNet simultaneously identifies tampered regions and their source counterparts. Extensive experiments on the benchmark bio-forensic datasets demonstrate significant improvements over competitive baselines in accurately detecting duplicated regions. Code - https://github.com/SoumyaroopNandi/BioTamperNet
Rescind: Countering Image Misconduct in Biomedical Publications with Vision-Language and State-Space Modeling
Jan 12, 2026Abstract:Scientific image manipulation in biomedical publications poses a growing threat to research integrity and reproducibility. Unlike natural image forensics, biomedical forgery detection is uniquely challenging due to domain-specific artifacts, complex textures, and unstructured figure layouts. We present the first vision-language guided framework for both generating and detecting biomedical image forgeries. By combining diffusion-based synthesis with vision-language prompting, our method enables realistic and semantically controlled manipulations, including duplication, splicing, and region removal, across diverse biomedical modalities. We introduce Rescind, a large-scale benchmark featuring fine-grained annotations and modality-specific splits, and propose Integscan, a structured state space modeling framework that integrates attention-enhanced visual encoding with prompt-conditioned semantic alignment for precise forgery localization. To ensure semantic fidelity, we incorporate a vision-language model based verification loop that filters generated forgeries based on consistency with intended prompts. Extensive experiments on Rescind and existing benchmarks demonstrate that Integscan achieves state of the art performance in both detection and localization, establishing a strong foundation for automated scientific integrity analysis.
TrainFors: A Large Benchmark Training Dataset for Image Manipulation Detection and Localization
Aug 10, 2023Abstract:The evaluation datasets and metrics for image manipulation detection and localization (IMDL) research have been standardized. But the training dataset for such a task is still nonstandard. Previous researchers have used unconventional and deviating datasets to train neural networks for detecting image forgeries and localizing pixel maps of manipulated regions. For a fair comparison, the training set, test set, and evaluation metrics should be persistent. Hence, comparing the existing methods may not seem fair as the results depend heavily on the training datasets as well as the model architecture. Moreover, none of the previous works release the synthetic training dataset used for the IMDL task. We propose a standardized benchmark training dataset for image splicing, copy-move forgery, removal forgery, and image enhancement forgery. Furthermore, we identify the problems with the existing IMDL datasets and propose the required modifications. We also train the state-of-the-art IMDL methods on our proposed TrainFors1 dataset for a fair evaluation and report the actual performance of these methods under similar conditions.
MONet: Multi-scale Overlap Network for Duplication Detection in Biomedical Images
Jul 19, 2022



Abstract:Manipulation of biomedical images to misrepresent experimental results has plagued the biomedical community for a while. Recent interest in the problem led to the curation of a dataset and associated tasks to promote the development of biomedical forensic methods. Of these, the largest manipulation detection task focuses on the detection of duplicated regions between images. Traditional computer-vision based forensic models trained on natural images are not designed to overcome the challenges presented by biomedical images. We propose a multi-scale overlap detection model to detect duplicated image regions. Our model is structured to find duplication hierarchically, so as to reduce the number of patch operations. It achieves state-of-the-art performance overall and on multiple biomedical image categories.
BioFors: A Large Biomedical Image Forensics Dataset
Aug 30, 2021



Abstract:Research in media forensics has gained traction to combat the spread of misinformation. However, most of this research has been directed towards content generated on social media. Biomedical image forensics is a related problem, where manipulation or misuse of images reported in biomedical research documents is of serious concern. The problem has failed to gain momentum beyond an academic discussion due to an absence of benchmark datasets and standardized tasks. In this paper we present BioFors -- the first dataset for benchmarking common biomedical image manipulations. BioFors comprises 47,805 images extracted from 1,031 open-source research papers. Images in BioFors are divided into four categories -- Microscopy, Blot/Gel, FACS and Macroscopy. We also propose three tasks for forensic analysis -- external duplication detection, internal duplication detection and cut/sharp-transition detection. We benchmark BioFors on all tasks with suitable state-of-the-art algorithms. Our results and analysis show that existing algorithms developed on common computer vision datasets are not robust when applied to biomedical images, validating that more research is required to address the unique challenges of biomedical image forensics.
SIGN: Spatial-information Incorporated Generative Network for Generalized Zero-shot Semantic Segmentation
Aug 27, 2021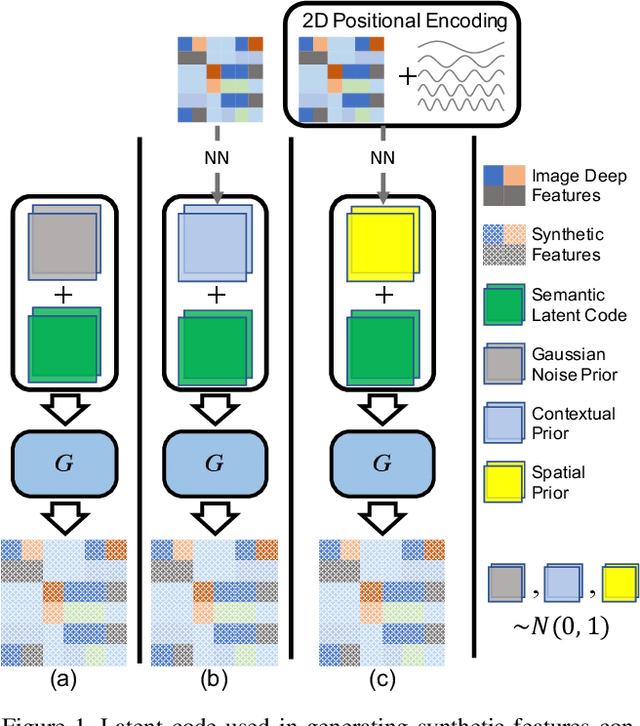
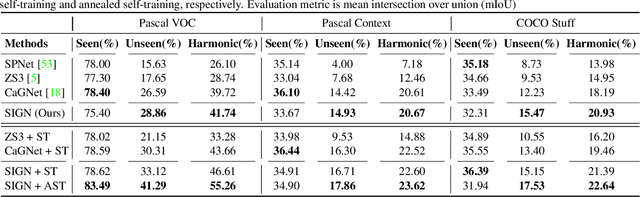
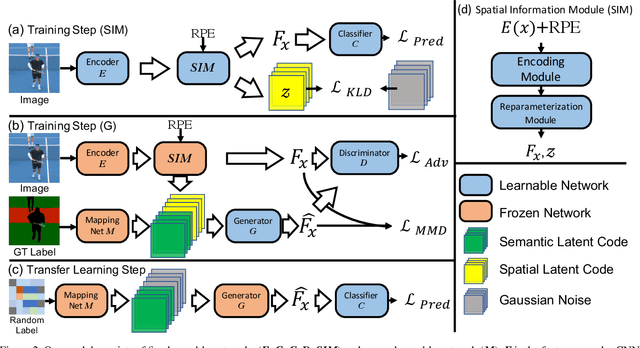
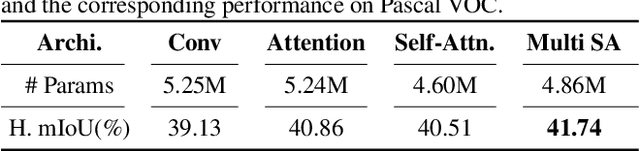
Abstract:Unlike conventional zero-shot classification, zero-shot semantic segmentation predicts a class label at the pixel level instead of the image level. When solving zero-shot semantic segmentation problems, the need for pixel-level prediction with surrounding context motivates us to incorporate spatial information using positional encoding. We improve standard positional encoding by introducing the concept of Relative Positional Encoding, which integrates spatial information at the feature level and can handle arbitrary image sizes. Furthermore, while self-training is widely used in zero-shot semantic segmentation to generate pseudo-labels, we propose a new knowledge-distillation-inspired self-training strategy, namely Annealed Self-Training, which can automatically assign different importance to pseudo-labels to improve performance. We systematically study the proposed Relative Positional Encoding and Annealed Self-Training in a comprehensive experimental evaluation, and our empirical results confirm the effectiveness of our method on three benchmark datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge