Soumen Ghosh
NeuroMorphix: A Novel Brain MRI Asymmetry-specific Feature Construction Approach For Seizure Recurrence Prediction
Apr 16, 2024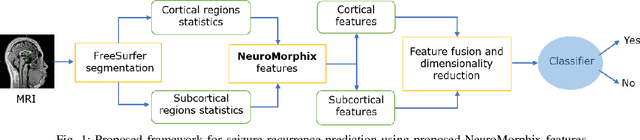
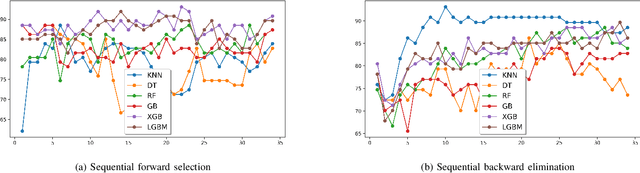

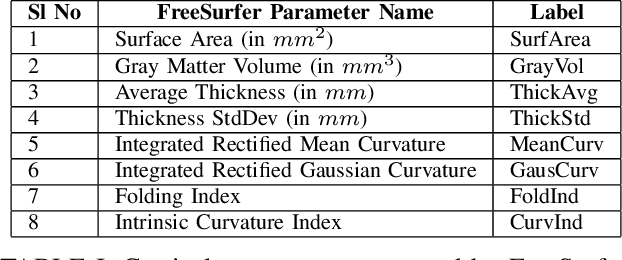
Abstract:Seizure recurrence is an important concern after an initial unprovoked seizure; without drug treatment, it occurs within 2 years in 40-50% of cases. The decision to treat currently relies on predictors of seizure recurrence risk that are inaccurate, resulting in unnecessary, possibly harmful, treatment in some patients and potentially preventable seizures in others. Because of the link between brain lesions and seizure recurrence, we developed a recurrence prediction tool using machine learning and clinical 3T brain MRI. We developed NeuroMorphix, a feature construction approach based on MRI brain anatomy. Each of seven NeuroMorphix features measures the absolute or relative difference between corresponding regions in each cerebral hemisphere. FreeSurfer was used to segment brain regions and to generate values for morphometric parameters (8 for each cortical region and 5 for each subcortical region). The parameters were then mapped to whole brain NeuroMorphix features, yielding a total of 91 features per subject. Features were generated for a first seizure patient cohort (n = 169) categorised into seizure recurrence and non-recurrence subgroups. State-of-the-art classification algorithms were trained and tested using NeuroMorphix features to predict seizure recurrence. Classification models using the top 5 features, ranked by sequential forward selection, demonstrated excellent performance in predicting seizure recurrence, with area under the ROC curve of 88-93%, accuracy of 83-89%, and F1 score of 83-90%. Highly ranked features aligned with structural alterations known to be associated with epilepsy. This study highlights the potential for targeted, data-driven approaches to aid clinical decision-making in brain disorders.
RCCNet: An Efficient Convolutional Neural Network for Histological Routine Colon Cancer Nuclei Classification
Oct 20, 2018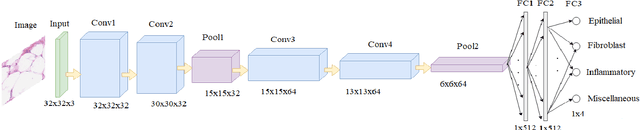
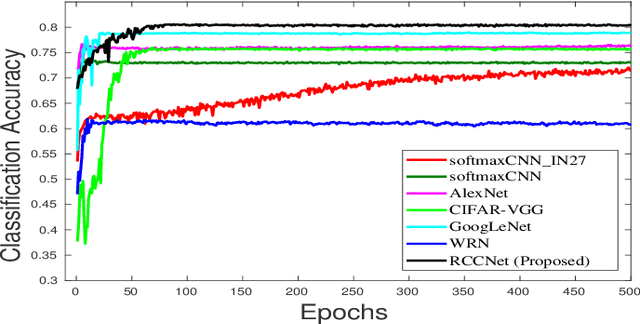
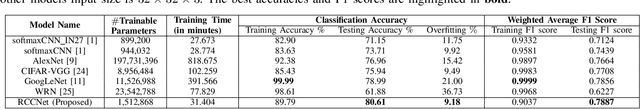
Abstract:Efficient and precise classification of histological cell nuclei is of utmost importance due to its potential applications in the field of medical image analysis. It would facilitate the medical practitioners to better understand and explore various factors for cancer treatment. The classification of histological cell nuclei is a challenging task due to the cellular heterogeneity. This paper proposes an efficient Convolutional Neural Network (CNN) based architecture for classification of histological routine colon cancer nuclei named as RCCNet. The main objective of this network is to keep the CNN model as simple as possible. The proposed RCCNet model consists of only 1,512,868 learnable parameters which are significantly less compared to the popular CNN models such as AlexNet, CIFARVGG, GoogLeNet, and WRN. The experiments are conducted over publicly available routine colon cancer histological dataset "CRCHistoPhenotypes". The results of the proposed RCCNet model are compared with five state-of-the-art CNN models in terms of the accuracy, weighted average F1 score and training time. The proposed method has achieved a classification accuracy of 80.61% and 0.7887 weighted average F1 score. The proposed RCCNet is more efficient and generalized terms of the training time and data over-fitting, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge