Sishuo Liu
RNC: Efficient RRAM-aware NAS and Compilation for DNNs on Resource-Constrained Edge Devices
Sep 27, 2024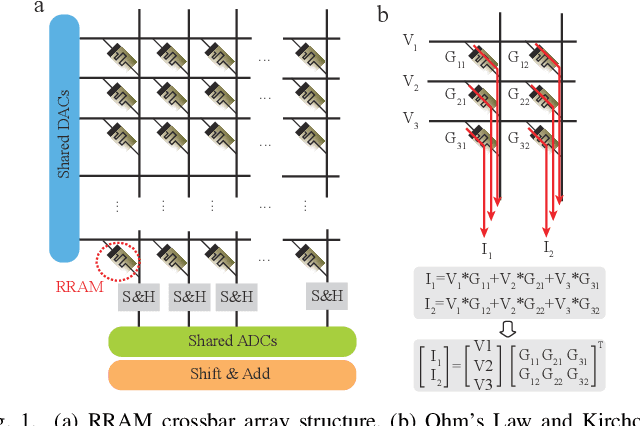
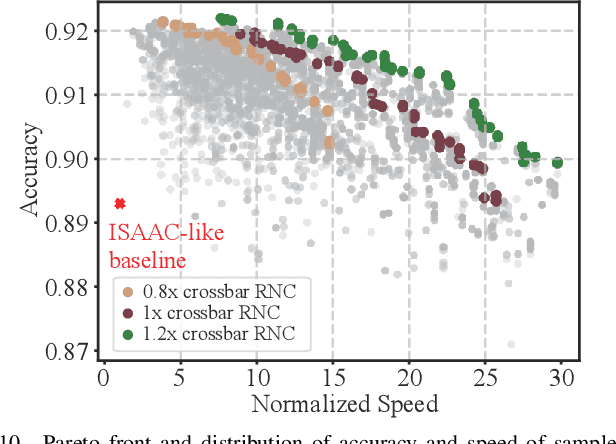
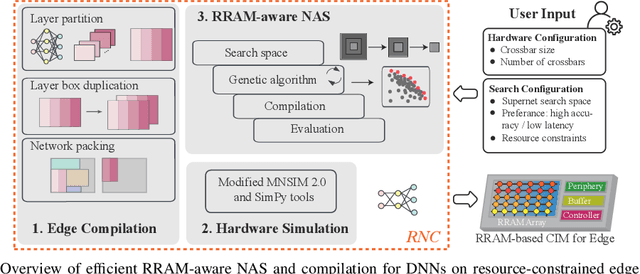
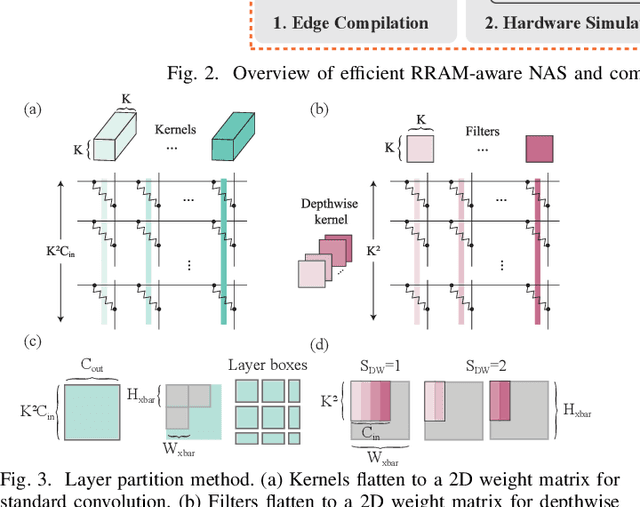
Abstract:Computing-in-memory (CIM) is an emerging computing paradigm, offering noteworthy potential for accelerating neural networks with high parallelism, low latency, and energy efficiency compared to conventional von Neumann architectures. However, existing research has primarily focused on hardware architecture and network co-design for large-scale neural networks, without considering resource constraints. In this study, we aim to develop edge-friendly deep neural networks (DNNs) for accelerators based on resistive random-access memory (RRAM). To achieve this, we propose an edge compilation and resource-constrained RRAM-aware neural architecture search (NAS) framework to search for optimized neural networks meeting specific hardware constraints. Our compilation approach integrates layer partitioning, duplication, and network packing to maximize the utilization of computation units. The resulting network architecture can be optimized for either high accuracy or low latency using a one-shot neural network approach with Pareto optimality achieved through the Non-dominated Sorted Genetic Algorithm II (NSGA-II). The compilation of mobile-friendly networks, like Squeezenet and MobilenetV3 small can achieve over 80% of utilization and over 6x speedup compared to ISAAC-like framework with different crossbar resources. The resulting model from NAS optimized for speed achieved 5x-30x speedup. The code for this paper is available at https://github.com/ArChiiii/rram_nas_comp_pack.
Self-Supervised Correction Learning for Semi-Supervised Biomedical Image Segmentation
Jan 12, 2023Abstract:Biomedical image segmentation plays a significant role in computer-aided diagnosis. However, existing CNN based methods rely heavily on massive manual annotations, which are very expensive and require huge human resources. In this work, we adopt a coarse-to-fine strategy and propose a self-supervised correction learning paradigm for semi-supervised biomedical image segmentation. Specifically, we design a dual-task network, including a shared encoder and two independent decoders for segmentation and lesion region inpainting, respectively. In the first phase, only the segmentation branch is used to obtain a relatively rough segmentation result. In the second step, we mask the detected lesion regions on the original image based on the initial segmentation map, and send it together with the original image into the network again to simultaneously perform inpainting and segmentation separately. For labeled data, this process is supervised by the segmentation annotations, and for unlabeled data, it is guided by the inpainting loss of masked lesion regions. Since the two tasks rely on similar feature information, the unlabeled data effectively enhances the representation of the network to the lesion regions and further improves the segmentation performance. Moreover, a gated feature fusion (GFF) module is designed to incorporate the complementary features from the two tasks. Experiments on three medical image segmentation datasets for different tasks including polyp, skin lesion and fundus optic disc segmentation well demonstrate the outstanding performance of our method compared with other semi-supervised approaches. The code is available at https://github.com/ReaFly/SemiMedSeg.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge