Siddhartha R. Jonnalagadda
An Information Extraction Approach to Prescreen Heart Failure Patients for Clinical Trials
Sep 06, 2016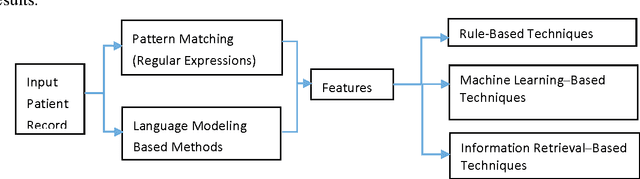
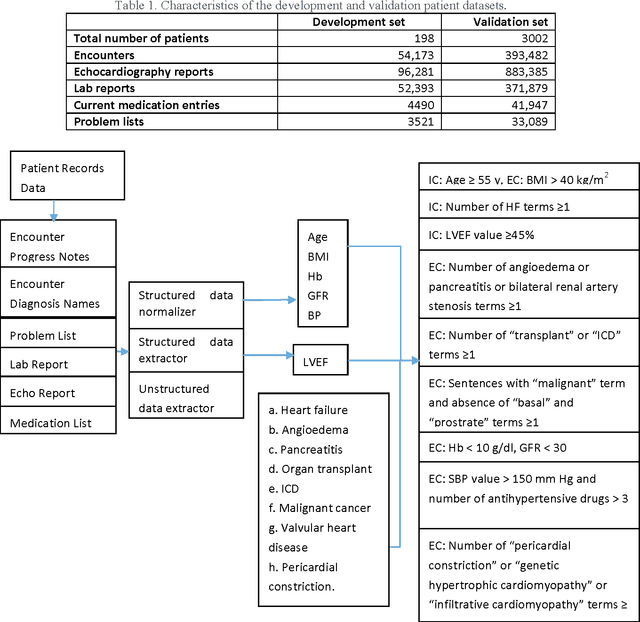
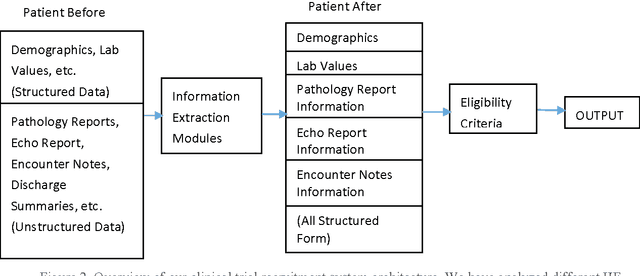
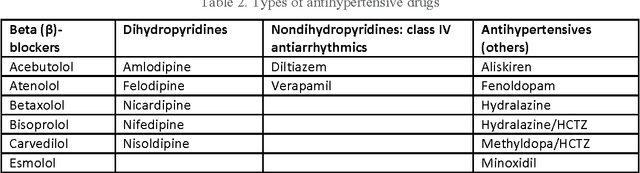
Abstract:To reduce the large amount of time spent screening, identifying, and recruiting patients into clinical trials, we need prescreening systems that are able to automate the data extraction and decision-making tasks that are typically relegated to clinical research study coordinators. However, a major obstacle is the vast amount of patient data available as unstructured free-form text in electronic health records. Here we propose an information extraction-based approach that first automatically converts unstructured text into a structured form. The structured data are then compared against a list of eligibility criteria using a rule-based system to determine which patients qualify for enrollment in a heart failure clinical trial. We show that we can achieve highly accurate results, with recall and precision values of 0.95 and 0.86, respectively. Our system allowed us to significantly reduce the time needed for prescreening patients from a few weeks to a few minutes. Our open-source information extraction modules are available for researchers and could be tested and validated in other cardiovascular trials. An approach such as the one we demonstrate here may decrease costs and expedite clinical trials, and could enhance the reproducibility of trials across institutions and populations.
Automatically extracting, ranking and visually summarizing the treatments for a disease
Sep 06, 2016

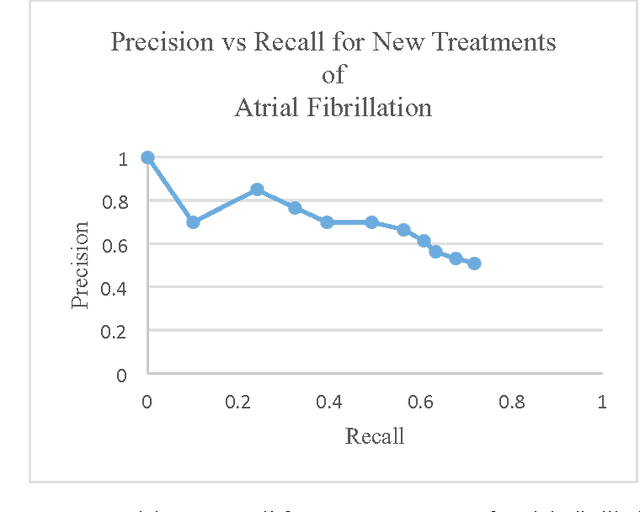

Abstract:Clinicians are expected to have up-to-date and broad knowledge of disease treatment options for a patient. Online health knowledge resources contain a wealth of information. However, because of the time investment needed to disseminate and rank pertinent information, there is a need to summarize the information in a more concise format. Our aim of the study is to provide clinicians with a concise overview of popular treatments for a given disease using information automatically computed from Medline abstracts. We analyzed the treatments of two disorders - Atrial Fibrillation and Congestive Heart Failure. We calculated the precision, recall, and f-scores of our two ranking methods to measure the accuracy of the results. For Atrial Fibrillation disorder, maximum f-score for the New Treatments weighing method is 0.611, which occurs at 60 treatments. For Congestive Heart Failure disorder, maximum f-score for the New Treatments weighing method is 0.503, which occurs at 80 treatments.
A Semi-supervised learning approach to enhance health care Community-based Question Answering: A case study in alcoholism
Jul 04, 2016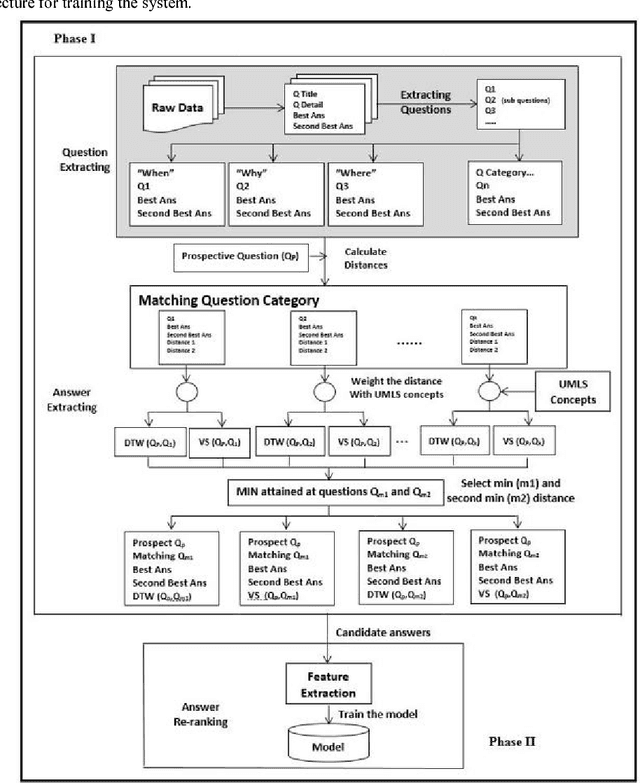
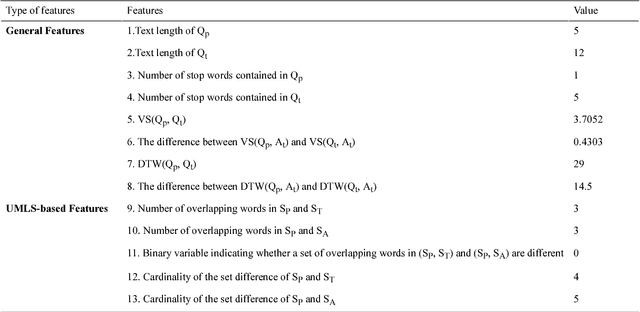
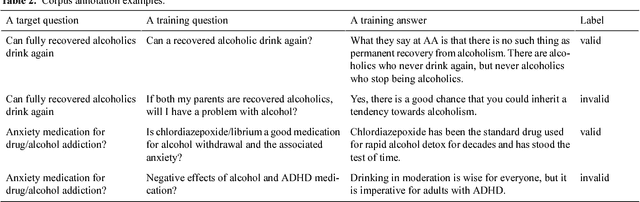
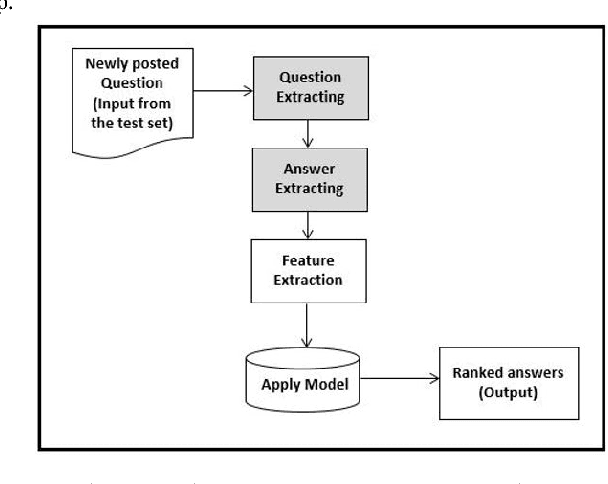
Abstract:Community-based Question Answering (CQA) sites play an important role in addressing health information needs. However, a significant number of posted questions remain unanswered. Automatically answering the posted questions can provide a useful source of information for online health communities. In this study, we developed an algorithm to automatically answer health-related questions based on past questions and answers (QA). We also aimed to understand information embedded within online health content that are good features in identifying valid answers. Our proposed algorithm uses information retrieval techniques to identify candidate answers from resolved QA. In order to rank these candidates, we implemented a semi-supervised leaning algorithm that extracts the best answer to a question. We assessed this approach on a curated corpus from Yahoo! Answers and compared against a rule-based string similarity baseline. On our dataset, the semi-supervised learning algorithm has an accuracy of 86.2%. UMLS-based (health-related) features used in the model enhance the algorithm's performance by proximately 8 %. A reasonably high rate of accuracy is obtained given that the data is considerably noisy. Important features distinguishing a valid answer from an invalid answer include text length, number of stop words contained in a test question, a distance between the test question and other questions in the corpus as well as a number of overlapping health-related terms between questions. Overall, our automated QA system based on historical QA pairs is shown to be effective according to the data set in this case study. It is developed for general use in the health care domain which can also be applied to other CQA sites.
A Novel Framework to Expedite Systematic Reviews by Automatically Building Information Extraction Training Corpora
Jun 21, 2016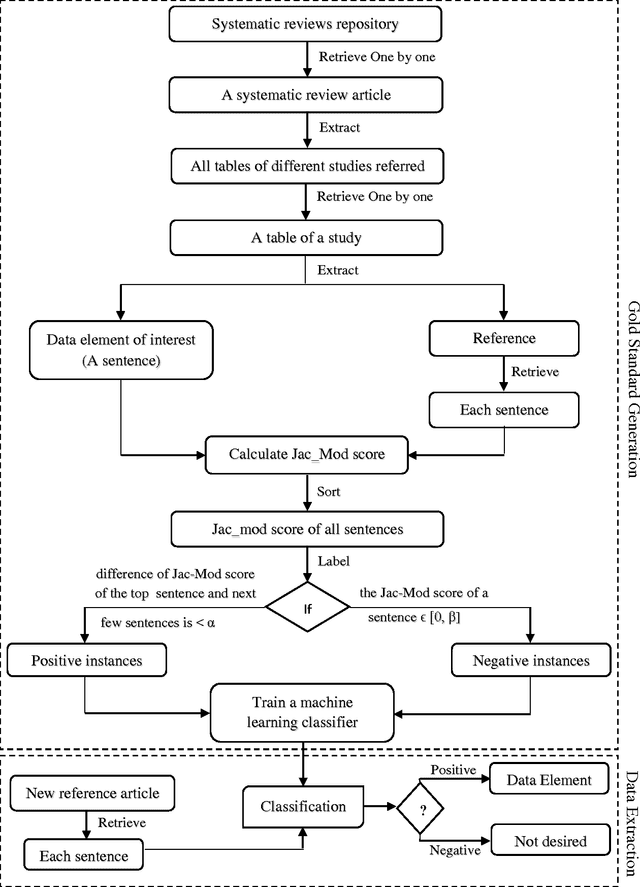
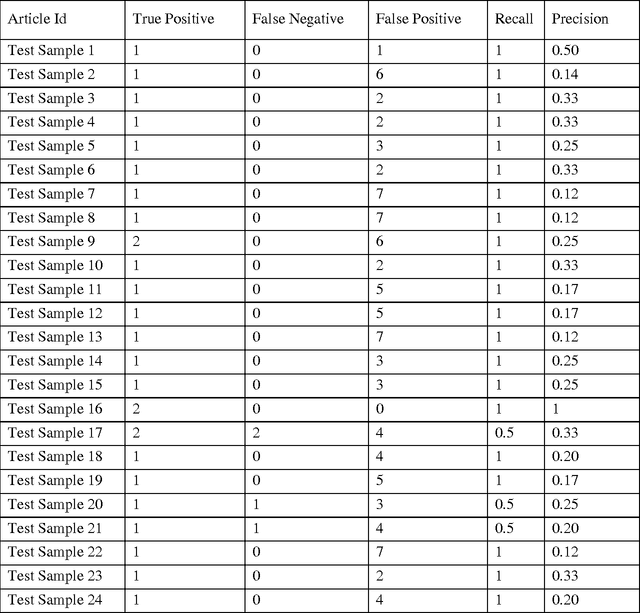
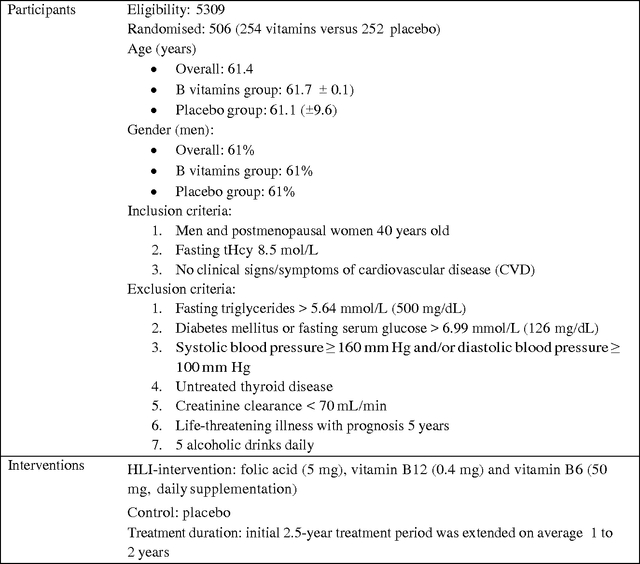
Abstract:A systematic review identifies and collates various clinical studies and compares data elements and results in order to provide an evidence based answer for a particular clinical question. The process is manual and involves lot of time. A tool to automate this process is lacking. The aim of this work is to develop a framework using natural language processing and machine learning to build information extraction algorithms to identify data elements in a new primary publication, without having to go through the expensive task of manual annotation to build gold standards for each data element type. The system is developed in two stages. Initially, it uses information contained in existing systematic reviews to identify the sentences from the PDF files of the included references that contain specific data elements of interest using a modified Jaccard similarity measure. These sentences have been treated as labeled data.A Support Vector Machine (SVM) classifier is trained on this labeled data to extract data elements of interests from a new article. We conducted experiments on Cochrane Database systematic reviews related to congestive heart failure using inclusion criteria as an example data element. The empirical results show that the proposed system automatically identifies sentences containing the data element of interest with a high recall (93.75%) and reasonable precision (27.05% - which means the reviewers have to read only 3.7 sentences on average). The empirical results suggest that the tool is retrieving valuable information from the reference articles, even when it is time-consuming to identify them manually. Thus we hope that the tool will be useful for automatic data extraction from biomedical research publications. The future scope of this work is to generalize this information framework for all types of systematic reviews.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge