Tanmay Basu
Identification of the Relevance of Comments in Codes Using Bag of Words and Transformer Based Models
Aug 11, 2023

Abstract:The Forum for Information Retrieval (FIRE) started a shared task this year for classification of comments of different code segments. This is binary text classification task where the objective is to identify whether comments given for certain code segments are relevant or not. The BioNLP-IISERB group at the Indian Institute of Science Education and Research Bhopal (IISERB) participated in this task and submitted five runs for five different models. The paper presents the overview of the models and other significant findings on the training corpus. The methods involve different feature engineering schemes and text classification techniques. The performance of the classical bag of words model and transformer-based models were explored to identify significant features from the given training corpus. We have explored different classifiers viz., random forest, support vector machine and logistic regression using the bag of words model. Furthermore, the pre-trained transformer based models like BERT, RoBERT and ALBERT were also used by fine-tuning them on the given training corpus. The performance of different such models over the training corpus were reported and the best five models were implemented on the given test corpus. The empirical results show that the bag of words model outperforms the transformer based models, however, the performance of our runs are not reasonably well in both training and test corpus. This paper also addresses the limitations of the models and scope for further improvement.
A Novel Framework to Expedite Systematic Reviews by Automatically Building Information Extraction Training Corpora
Jun 21, 2016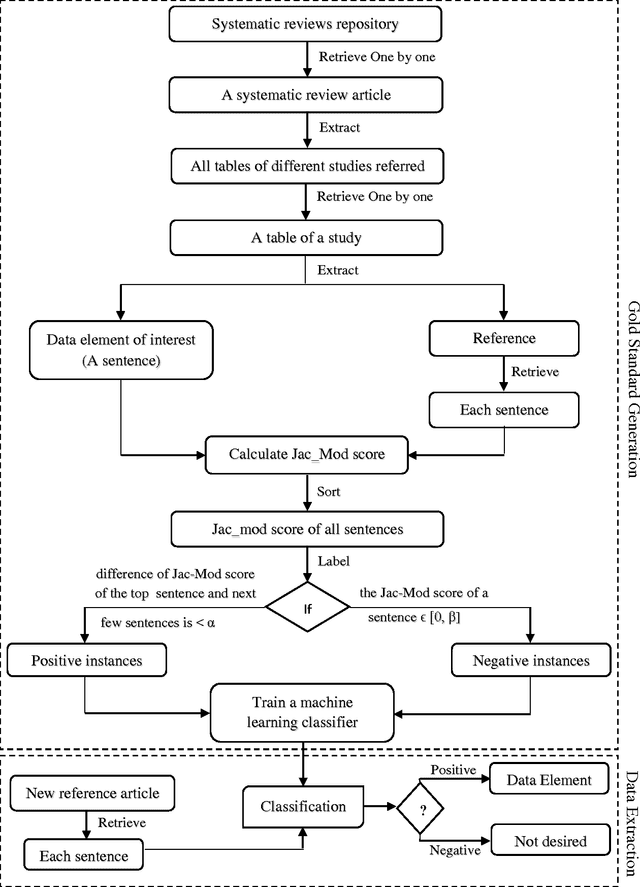
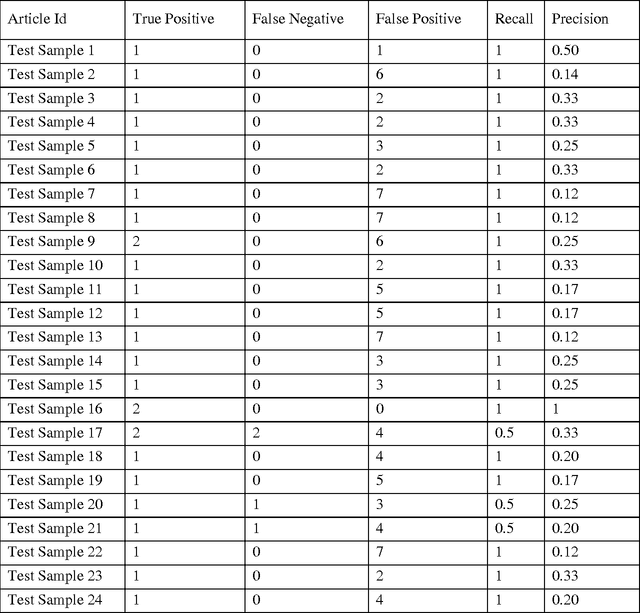
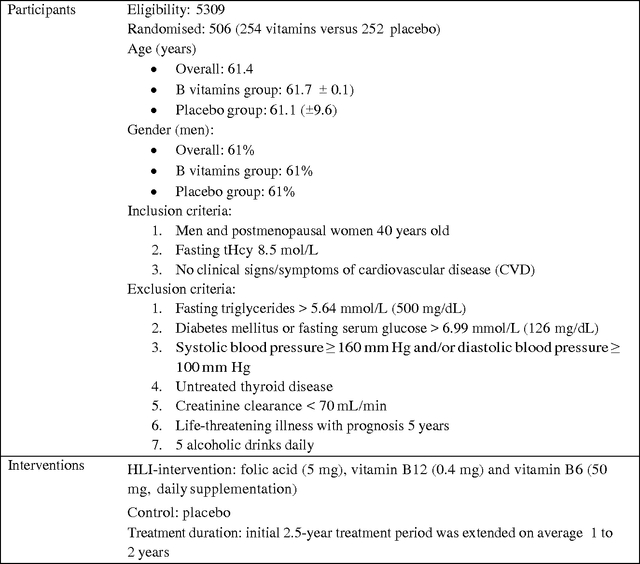
Abstract:A systematic review identifies and collates various clinical studies and compares data elements and results in order to provide an evidence based answer for a particular clinical question. The process is manual and involves lot of time. A tool to automate this process is lacking. The aim of this work is to develop a framework using natural language processing and machine learning to build information extraction algorithms to identify data elements in a new primary publication, without having to go through the expensive task of manual annotation to build gold standards for each data element type. The system is developed in two stages. Initially, it uses information contained in existing systematic reviews to identify the sentences from the PDF files of the included references that contain specific data elements of interest using a modified Jaccard similarity measure. These sentences have been treated as labeled data.A Support Vector Machine (SVM) classifier is trained on this labeled data to extract data elements of interests from a new article. We conducted experiments on Cochrane Database systematic reviews related to congestive heart failure using inclusion criteria as an example data element. The empirical results show that the proposed system automatically identifies sentences containing the data element of interest with a high recall (93.75%) and reasonable precision (27.05% - which means the reviewers have to read only 3.7 sentences on average). The empirical results suggest that the tool is retrieving valuable information from the reference articles, even when it is time-consuming to identify them manually. Thus we hope that the tool will be useful for automatic data extraction from biomedical research publications. The future scope of this work is to generalize this information framework for all types of systematic reviews.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge