Shir Bar
Sifting through the haystack -- efficiently finding rare animal behaviors in large-scale datasets
Dec 04, 2024Abstract:In the study of animal behavior, researchers often record long continuous videos, accumulating into large-scale datasets. However, the behaviors of interest are often rare compared to routine behaviors. This incurs a heavy cost on manual annotation, forcing users to sift through many samples before finding their needles. We propose a pipeline to efficiently sample rare behaviors from large datasets, enabling the creation of training datasets for rare behavior classifiers. Our method only needs an unlabeled animal pose or acceleration dataset as input and makes no assumptions regarding the type, number, or characteristics of the rare behaviors. Our pipeline is based on a recent graph-based anomaly detection model for human behavior, which we apply to this new data domain. It leverages anomaly scores to automatically label normal samples while directing human annotation efforts toward anomalies. In research data, anomalies may come from many different sources (e.g., signal noise versus true rare instances). Hence, the entire labeling budget is focused on the abnormal classes, letting the user review and label samples according to their needs. We tested our approach on three datasets of freely-moving animals, acquired in the laboratory and the field. We found that graph-based models are particularly useful when studying motion-based behaviors in animals, yielding good results while using a small labeling budget. Our method consistently outperformed traditional random sampling, offering an average improvement of 70% in performance and creating datasets even when the behavior of interest was only 0.02% of the data. Even when the performance gain was minor (e.g., when the behavior is not rare), our method still reduced the annotation effort by half
TextRay: Mining Clinical Reports to Gain a Broad Understanding of Chest X-rays
Jun 06, 2018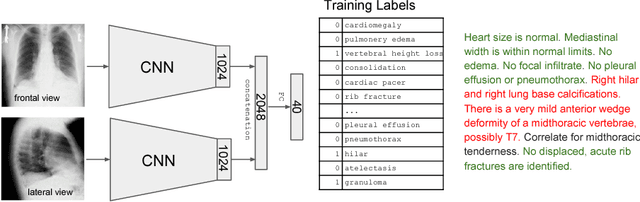
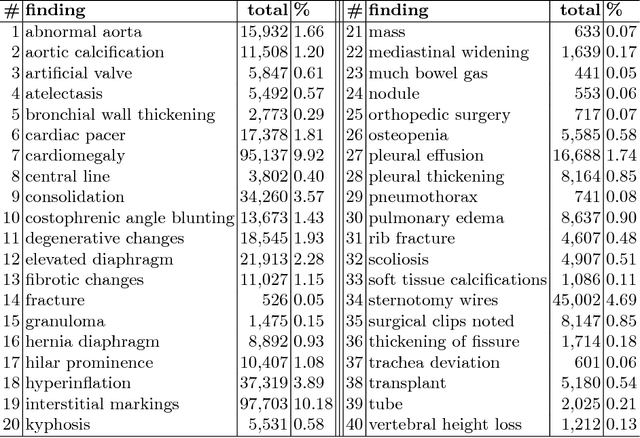
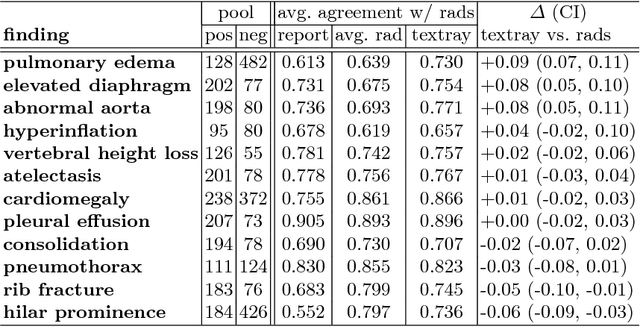
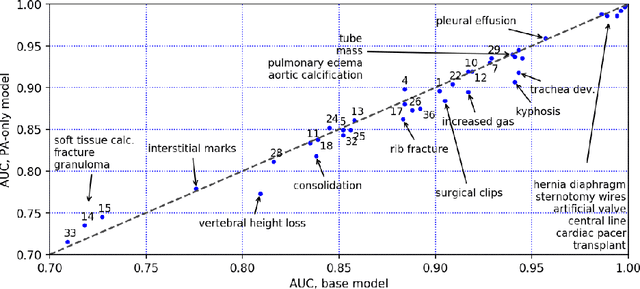
Abstract:The chest X-ray (CXR) is by far the most commonly performed radiological examination for screening and diagnosis of many cardiac and pulmonary diseases. There is an immense world-wide shortage of physicians capable of providing rapid and accurate interpretation of this study. A radiologist-driven analysis of over two million CXR reports generated an ontology including the 40 most prevalent pathologies on CXR. By manually tagging a relatively small set of sentences, we were able to construct a training set of 959k studies. A deep learning model was trained to predict the findings given the patient frontal and lateral scans. For 12 of the findings we compare the model performance against a team of radiologists and show that in most cases the radiologists agree on average more with the algorithm than with each other.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge