Shengda Liu
KrwEmd: Revising the Imperfect-Recall Abstraction from Forgetting Everything
Nov 15, 2025Abstract:Excessive abstraction is a critical challenge in hand abstraction-a task specific to games like Texas hold'em-when solving large-scale imperfect-information games, as it impairs AI performance. This issue arises from extreme implementations of imperfect-recall abstraction, which entirely discard historical information. This paper presents KrwEmd, the first practical algorithm designed to address this problem. We first introduce the k-recall winrate feature, which not only qualitatively distinguishes signal observation infosets by leveraging both future and, crucially, historical game information, but also quantitatively captures their similarity. We then develop the KrwEmd algorithm, which clusters signal observation infosets using earth mover's distance to measure discrepancies between their features. Experimental results demonstrate that KrwEmd significantly improves AI gameplay performance compared to existing algorithms.
No-Regret Strategy Solving in Imperfect-Information Games via Pre-Trained Embedding
Nov 15, 2025Abstract:High-quality information set abstraction remains a core challenge in solving large-scale imperfect-information extensive-form games (IIEFGs)-such as no-limit Texas Hold'em-where the finite nature of spatial resources hinders strategy solving over the full game. State-of-the-art AI methods rely on pre-trained discrete clustering for abstraction, yet their hard classification irreversibly loses critical information: specifically, the quantifiable subtle differences between information sets-vital for strategy solving-thereby compromising the quality of such solving. Inspired by the word embedding paradigm in natural language processing, this paper proposes the Embedding CFR algorithm, a novel approach for solving strategies in IIEFGs within an embedding space. The algorithm pre-trains and embeds features of isolated information sets into an interconnected low-dimensional continuous space, where the resulting vectors more precisely capture both the distinctions and connections between information sets. Embedding CFR presents a strategy-solving process driven by regret accumulation and strategy updates within this embedding space, with accompanying theoretical analysis verifying its capacity to reduce cumulative regret. Experiments on poker show that with the same spatial overhead, Embedding CFR achieves significantly faster exploitability convergence compared to cluster-based abstraction algorithms, confirming its effectiveness. Furthermore, to our knowledge, it is the first algorithm in poker AI that pre-trains information set abstractions through low-dimensional embedding for strategy solving.
WGSR-Bench: Wargame-based Game-theoretic Strategic Reasoning Benchmark for Large Language Models
Jun 12, 2025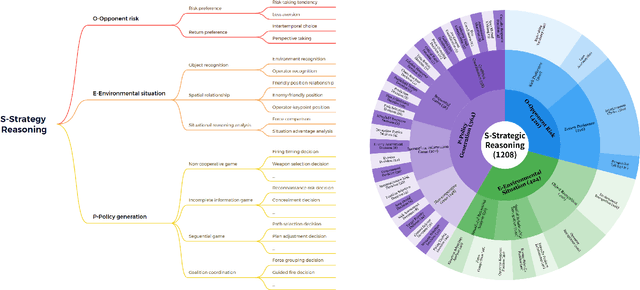
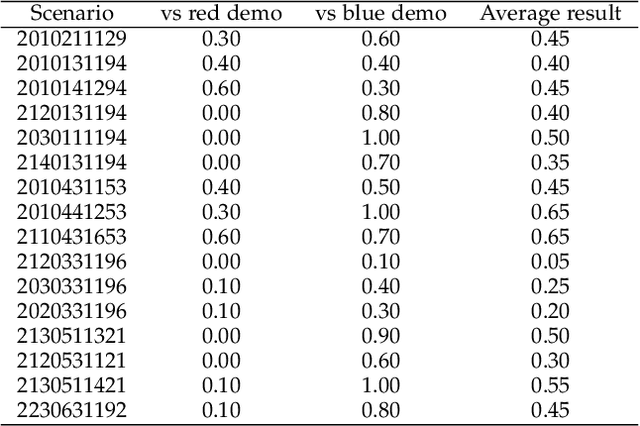
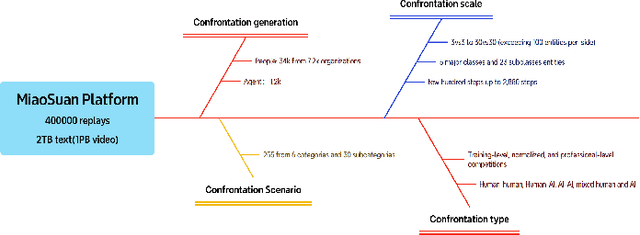
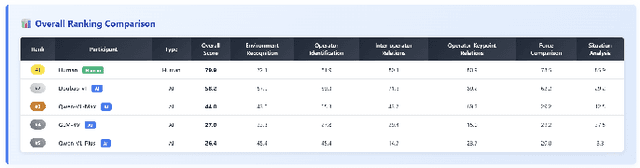
Abstract:Recent breakthroughs in Large Language Models (LLMs) have led to a qualitative leap in artificial intelligence' s performance on reasoning tasks, particularly demonstrating remarkable capabilities in mathematical, symbolic, and commonsense reasoning. However, as a critical component of advanced human cognition, strategic reasoning, i.e., the ability to assess multi-agent behaviors in dynamic environments, formulate action plans, and adapt strategies, has yet to be systematically evaluated or modeled. To address this gap, this paper introduces WGSR-Bench, the first strategy reasoning benchmark for LLMs using wargame as its evaluation environment. Wargame, a quintessential high-complexity strategic scenario, integrates environmental uncertainty, adversarial dynamics, and non-unique strategic choices, making it an effective testbed for assessing LLMs' capabilities in multi-agent decision-making, intent inference, and counterfactual reasoning. WGSR-Bench designs test samples around three core tasks, i.e., Environmental situation awareness, Opponent risk modeling and Policy generation, which serve as the core S-POE architecture, to systematically assess main abilities of strategic reasoning. Finally, an LLM-based wargame agent is designed to integrate these parts for a comprehensive strategy reasoning assessment. With WGSR-Bench, we hope to assess the strengths and limitations of state-of-the-art LLMs in game-theoretic strategic reasoning and to advance research in large model-driven strategic intelligence.
Imbalance-Aware Self-Supervised Learning for 3D Radiomic Representations
Mar 06, 2021



Abstract:Radiomic representations can quantify properties of regions of interest in medical image data. Classically, they account for pre-defined statistics of shape, texture, and other low-level image features. Alternatively, deep learning-based representations are derived from supervised learning but require expensive annotations from experts and often suffer from overfitting and data imbalance issues. In this work, we address the challenge of learning representations of 3D medical images for an effective quantification under data imbalance. We propose a \emph{self-supervised} representation learning framework to learn high-level features of 3D volumes as a complement to existing radiomics features. Specifically, we demonstrate how to learn image representations in a self-supervised fashion using a 3D Siamese network. More importantly, we deal with data imbalance by exploiting two unsupervised strategies: a) sample re-weighting, and b) balancing the composition of training batches. When combining our learned self-supervised feature with traditional radiomics, we show significant improvement in brain tumor classification and lung cancer staging tasks covering MRI and CT imaging modalities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge