Seungheun Baek
ATTNSOM: Learning Cross-Isoform Attention for Cytochrome P450 Site-of-Metabolism
Jan 28, 2026Abstract:Identifying metabolic sites where cytochrome P450 enzymes metabolize small-molecule drugs is essential for drug discovery. Although existing computational approaches have been proposed for site-of-metabolism prediction, they typically ignore cytochrome P450 isoform identity or model isoforms independently, thereby failing to fully capture inherent cross-isoform metabolic patterns. In addition, prior evaluations often rely on top-k metrics, where false positive atoms may be included among the top predictions, underscoring the need for complementary metrics that more directly assess binary atom-level discrimination under severe class imbalance. We propose ATTNSOM, an atom-level site-of-metabolism prediction framework that integrates intrinsic molecular reactivity with cross-isoform relationships. The model combines a shared graph encoder, molecule-conditioned atom representations, and a cross-attention mechanism to capture correlated metabolic patterns across cytochrome P450 isoforms. The model is evaluated on two benchmark datasets annotated with site-of-metabolism labels at atom resolution. Across these benchmarks, the model achieves consistently strong top-k performance across multiple cytochrome P450 isoforms. Relative to ablated variants, the model yields higher Matthews correlation coefficient, indicating improved discrimination of true metabolic sites. These results support the importance of explicitly modeling cross-isoform relationships for site-of-metabolism prediction. The code and datasets are available at https://github.com/dmis-lab/ATTNSOM.
HiRef: Leveraging Hierarchical Ontology and Network Refinement for Robust Medication Recommendation
Aug 14, 2025Abstract:Medication recommendation is a crucial task for assisting physicians in making timely decisions from longitudinal patient medical records. However, real-world EHR data present significant challenges due to the presence of rarely observed medical entities and incomplete records that may not fully capture the clinical ground truth. While data-driven models trained on longitudinal Electronic Health Records often achieve strong empirical performance, they struggle to generalize under missing or novel conditions, largely due to their reliance on observed co-occurrence patterns. To address these issues, we propose Hierarchical Ontology and Network Refinement for Robust Medication Recommendation (HiRef), a unified framework that combines two complementary structures: (i) the hierarchical semantics encoded in curated medical ontologies, and (ii) refined co-occurrence patterns derived from real-world EHRs. We embed ontology entities in hyperbolic space, which naturally captures tree-like relationships and enables knowledge transfer through shared ancestors, thereby improving generalizability to unseen codes. To further improve robustness, we introduce a prior-guided sparse regularization scheme that refines the EHR co-occurrence graph by suppressing spurious edges while preserving clinically meaningful associations. Our model achieves strong performance on EHR benchmarks (MIMIC-III and MIMIC-IV) and maintains high accuracy under simulated unseen-code settings. Extensive experiments with comprehensive ablation studies demonstrate HiRef's resilience to unseen medical codes, supported by in-depth analyses of the learned sparsified graph structure and medical code embeddings.
GPO-VAE: Modeling Explainable Gene Perturbation Responses utilizing GRN-Aligned Parameter Optimization
Jan 31, 2025Abstract:Motivation: Predicting cellular responses to genetic perturbations is essential for understanding biological systems and developing targeted therapeutic strategies. While variational autoencoders (VAEs) have shown promise in modeling perturbation responses, their limited explainability poses a significant challenge, as the learned features often lack clear biological meaning. Nevertheless, model explainability is one of the most important aspects in the realm of biological AI. One of the most effective ways to achieve explainability is incorporating the concept of gene regulatory networks (GRNs) in designing deep learning models such as VAEs. GRNs elicit the underlying causal relationships between genes and are capable of explaining the transcriptional responses caused by genetic perturbation treatments. Results: We propose GPO-VAE, an explainable VAE enhanced by GRN-aligned Parameter Optimization that explicitly models gene regulatory networks in the latent space. Our key approach is to optimize the learnable parameters related to latent perturbation effects towards GRN-aligned explainability. Experimental results on perturbation prediction show our model achieves state-of-the-art performance in predicting transcriptional responses across multiple benchmark datasets. Furthermore, additional results on evaluating the GRN inference task reveal our model's ability to generate meaningful GRNs compared to other methods. According to qualitative analysis, GPO-VAE posseses the ability to construct biologically explainable GRNs that align with experimentally validated regulatory pathways. GPO-VAE is available at https://github.com/dmis-lab/GPO-VAE
CRADLE-VAE: Enhancing Single-Cell Gene Perturbation Modeling with Counterfactual Reasoning-based Artifact Disentanglement
Sep 10, 2024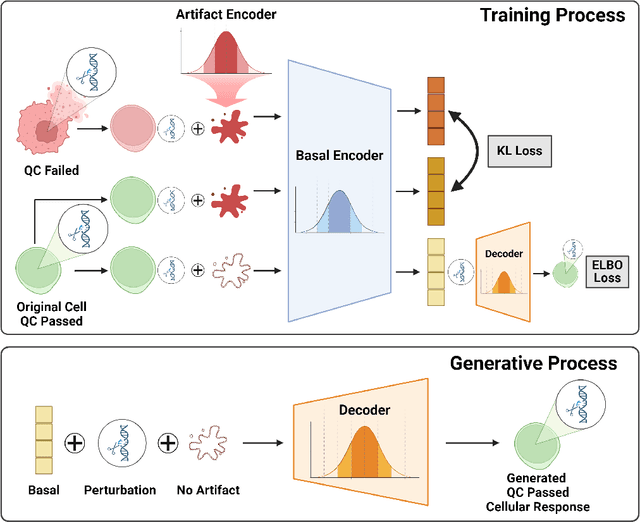
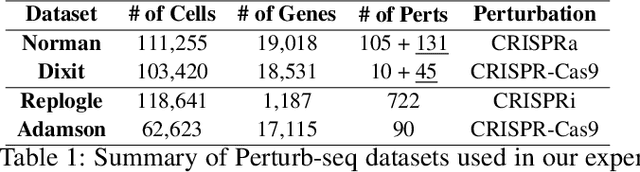
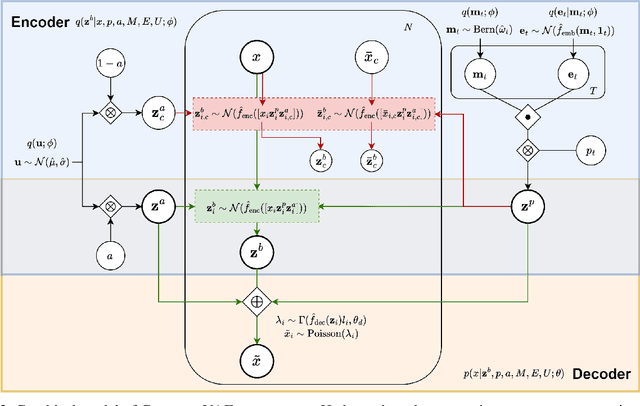
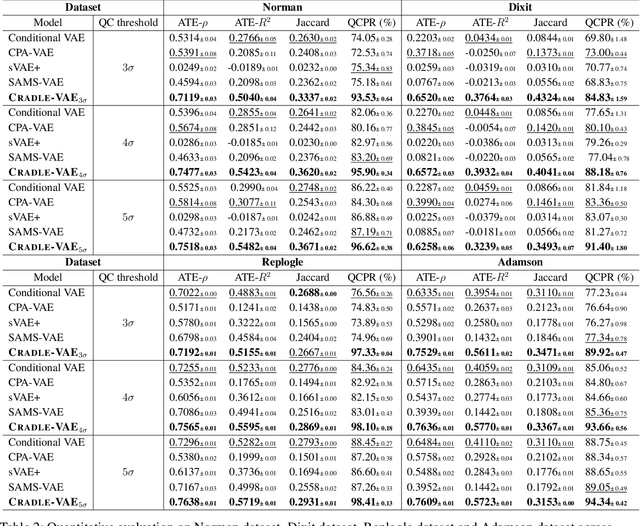
Abstract:Predicting cellular responses to various perturbations is a critical focus in drug discovery and personalized therapeutics, with deep learning models playing a significant role in this endeavor. Single-cell datasets contain technical artifacts that may hinder the predictability of such models, which poses quality control issues highly regarded in this area. To address this, we propose CRADLE-VAE, a causal generative framework tailored for single-cell gene perturbation modeling, enhanced with counterfactual reasoning-based artifact disentanglement. Throughout training, CRADLE-VAE models the underlying latent distribution of technical artifacts and perturbation effects present in single-cell datasets. It employs counterfactual reasoning to effectively disentangle such artifacts by modulating the latent basal spaces and learns robust features for generating cellular response data with improved quality. Experimental results demonstrate that this approach improves not only treatment effect estimation performance but also generative quality as well. The CRADLE-VAE codebase is publicly available at https://github.com/dmis-lab/CRADLE-VAE.
MolPLA: A Molecular Pretraining Framework for Learning Cores, R-Groups and their Linker Joints
Jan 30, 2024Abstract:Molecular core structures and R-groups are essential concepts in drug development. Integration of these concepts with conventional graph pre-training approaches can promote deeper understanding in molecules. We propose MolPLA, a novel pre-training framework that employs masked graph contrastive learning in understanding the underlying decomposable parts inmolecules that implicate their core structure and peripheral R-groups. Furthermore, we formulate an additional framework that grants MolPLA the ability to help chemists find replaceable R-groups in lead optimization scenarios. Experimental results on molecular property prediction show that MolPLA exhibits predictability comparable to current state-of-the-art models. Qualitative analysis implicate that MolPLA is capable of distinguishing core and R-group sub-structures, identifying decomposable regions in molecules and contributing to lead optimization scenarios by rationally suggesting R-group replacements given various query core templates. The code implementation for MolPLA and its pre-trained model checkpoint is available at https://github.com/dmis-lab/MolPLA
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge