Sayed Hashim
Self-omics: A Self-supervised Learning Framework for Multi-omics Cancer Data
Oct 03, 2022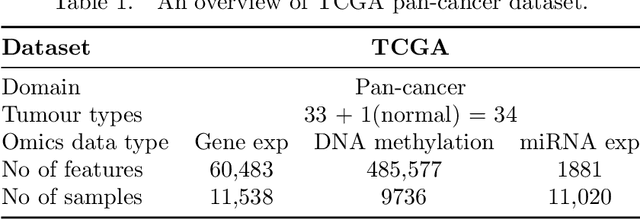
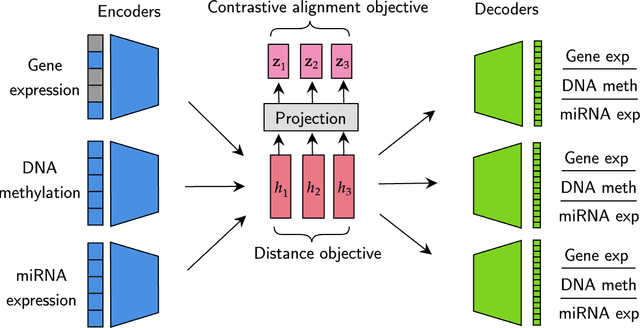
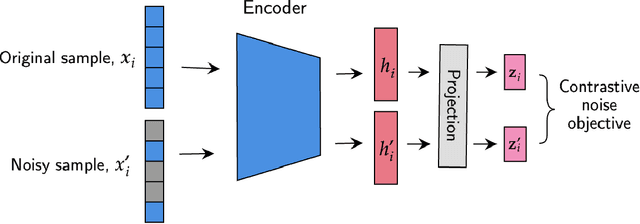
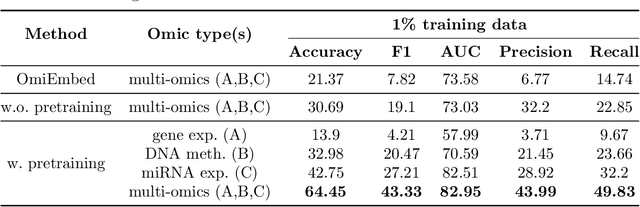
Abstract:We have gained access to vast amounts of multi-omics data thanks to Next Generation Sequencing. However, it is challenging to analyse this data due to its high dimensionality and much of it not being annotated. Lack of annotated data is a significant problem in machine learning, and Self-Supervised Learning (SSL) methods are typically used to deal with limited labelled data. However, there is a lack of studies that use SSL methods to exploit inter-omics relationships on unlabelled multi-omics data. In this work, we develop a novel and efficient pre-training paradigm that consists of various SSL components, including but not limited to contrastive alignment, data recovery from corrupted samples, and using one type of omics data to recover other omic types. Our pre-training paradigm improves performance on downstream tasks with limited labelled data. We show that our approach outperforms the state-of-the-art method in cancer type classification on the TCGA pan-cancer dataset in semi-supervised setting. Moreover, we show that the encoders that are pre-trained using our approach can be used as powerful feature extractors even without fine-tuning. Our ablation study shows that the method is not overly dependent on any pretext task component. The network architectures in our approach are designed to handle missing omic types and multiple datasets for pre-training and downstream training. Our pre-training paradigm can be extended to perform zero-shot classification of rare cancers.
Survey on Self-supervised Representation Learning Using Image Transformations
Feb 17, 2022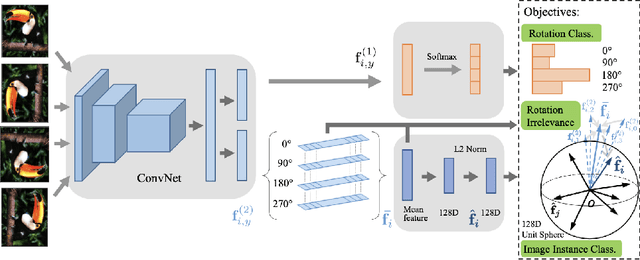
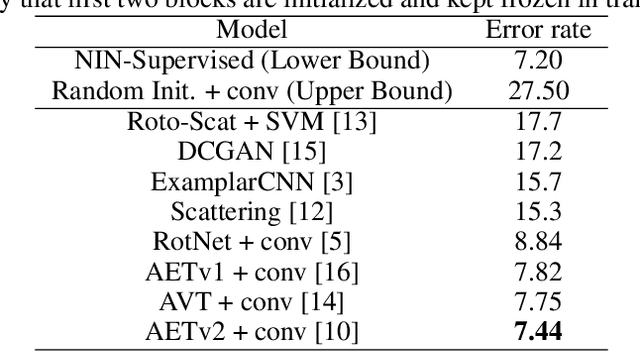
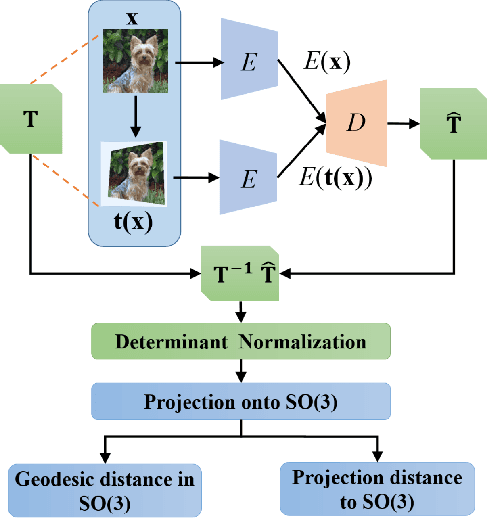
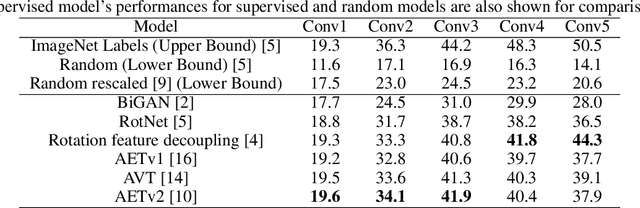
Abstract:Deep neural networks need huge amount of training data, while in real world there is a scarcity of data available for training purposes. To resolve these issues, self-supervised learning (SSL) methods are used. SSL using geometric transformations (GT) is a simple yet powerful technique used in unsupervised representation learning. Although multiple survey papers have reviewed SSL techniques, there is none that only focuses on those that use geometric transformations. Furthermore, such methods have not been covered in depth in papers where they are reviewed. Our motivation to present this work is that geometric transformations have shown to be powerful supervisory signals in unsupervised representation learning. Moreover, many such works have found tremendous success, but have not gained much attention. We present a concise survey of SSL approaches that use geometric transformations. We shortlist six representative models that use image transformations including those based on predicting and autoencoding transformations. We review their architecture as well as learning methodologies. We also compare the performance of these models in the object recognition task on CIFAR-10 and ImageNet datasets. Our analysis indicates the AETv2 performs the best in most settings. Rotation with feature decoupling also performed well in some settings. We then derive insights from the observed results. Finally, we conclude with a summary of the results and insights as well as highlighting open problems to be addressed and indicating various future directions.
TransformNet: Self-supervised representation learning through predicting geometric transformations
Feb 08, 2022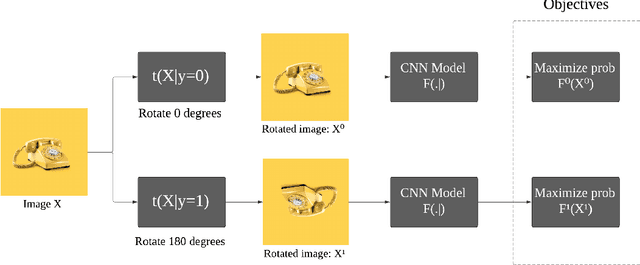
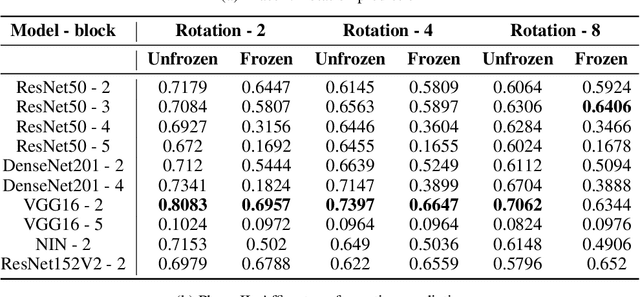
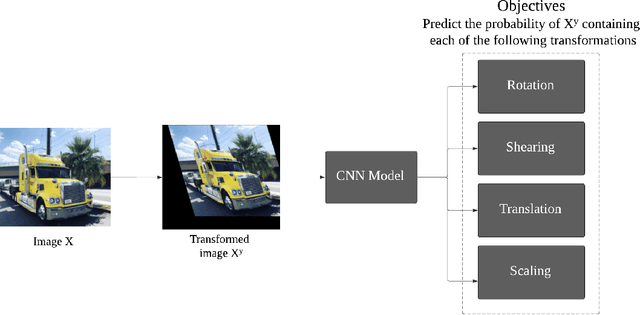

Abstract:Deep neural networks need a big amount of training data, while in the real world there is a scarcity of data available for training purposes. To resolve this issue unsupervised methods are used for training with limited data. In this report, we describe the unsupervised semantic feature learning approach for recognition of the geometric transformation applied to the input data. The basic concept of our approach is that if someone is unaware of the objects in the images, he/she would not be able to quantitatively predict the geometric transformation that was applied to them. This self supervised scheme is based on pretext task and the downstream task. The pretext classification task to quantify the geometric transformations should force the CNN to learn high-level salient features of objects useful for image classification. In our baseline model, we define image rotations by multiples of 90 degrees. The CNN trained on this pretext task will be used for the classification of images in the CIFAR-10 dataset as a downstream task. we run the baseline method using various models, including ResNet, DenseNet, VGG-16, and NIN with a varied number of rotations in feature extracting and fine-tuning settings. In extension of this baseline model we experiment with transformations other than rotation in pretext task. We compare performance of selected models in various settings with different transformations applied to images,various data augmentation techniques as well as using different optimizers. This series of different type of experiments will help us demonstrate the recognition accuracy of our self-supervised model when applied to a downstream task of classification.
SubOmiEmbed: Self-supervised Representation Learning of Multi-omics Data for Cancer Type Classification
Feb 03, 2022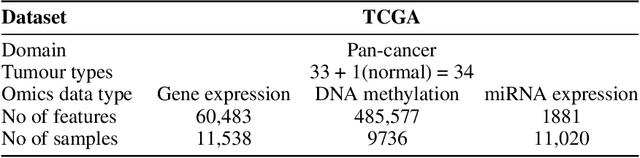
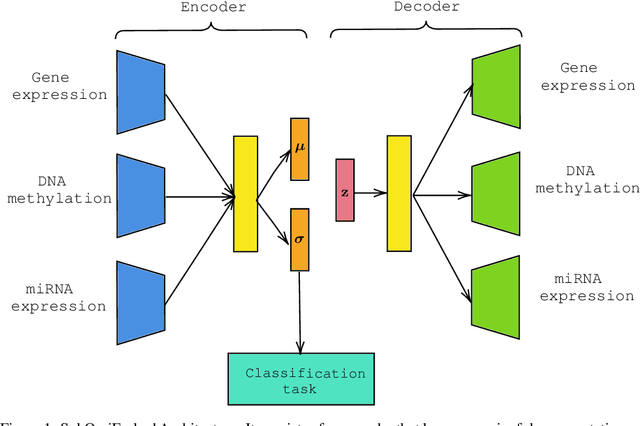
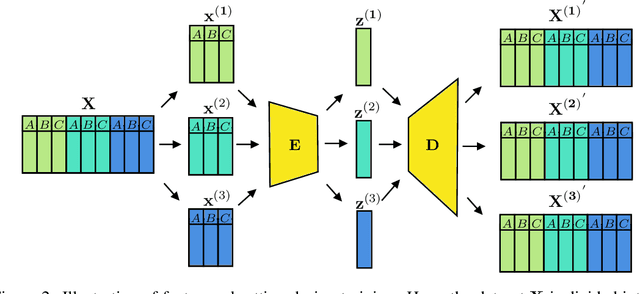
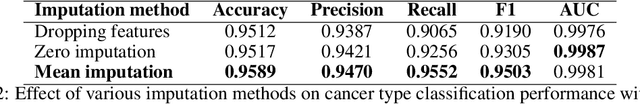
Abstract:For personalized medicines, very crucial intrinsic information is present in high dimensional omics data which is difficult to capture due to the large number of molecular features and small number of available samples. Different types of omics data show various aspects of samples. Integration and analysis of multi-omics data give us a broad view of tumours, which can improve clinical decision making. Omics data, mainly DNA methylation and gene expression profiles are usually high dimensional data with a lot of molecular features. In recent years, variational autoencoders (VAE) have been extensively used in embedding image and text data into lower dimensional latent spaces. In our project, we extend the idea of using a VAE model for low dimensional latent space extraction with the self-supervised learning technique of feature subsetting. With VAEs, the key idea is to make the model learn meaningful representations from different types of omics data, which could then be used for downstream tasks such as cancer type classification. The main goals are to overcome the curse of dimensionality and integrate methylation and expression data to combine information about different aspects of same tissue samples, and hopefully extract biologically relevant features. Our extension involves training encoder and decoder to reconstruct the data from just a subset of it. By doing this, we force the model to encode most important information in the latent representation. We also added an identity to the subsets so that the model knows which subset is being fed into it during training and testing. We experimented with our approach and found that SubOmiEmbed produces comparable results to the baseline OmiEmbed with a much smaller network and by using just a subset of the data. This work can be improved to integrate mutation-based genomic data as well.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge