SubOmiEmbed: Self-supervised Representation Learning of Multi-omics Data for Cancer Type Classification
Paper and Code
Feb 03, 2022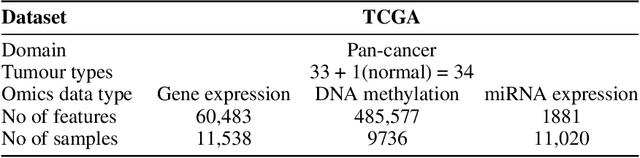
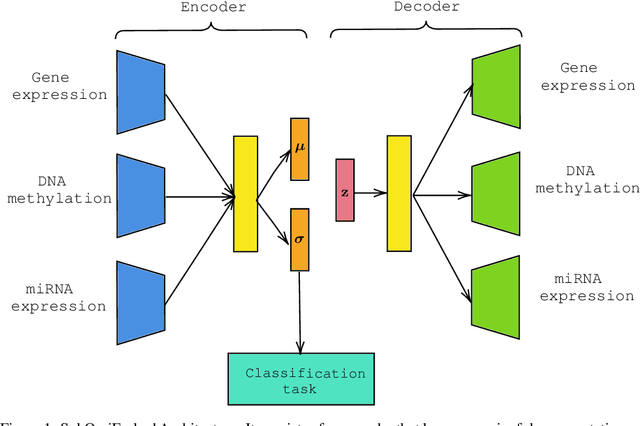
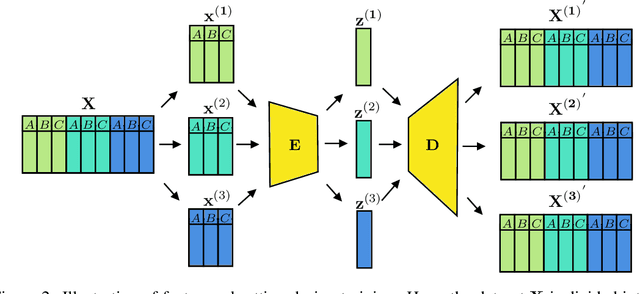
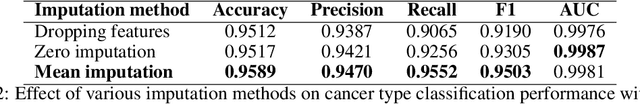
For personalized medicines, very crucial intrinsic information is present in high dimensional omics data which is difficult to capture due to the large number of molecular features and small number of available samples. Different types of omics data show various aspects of samples. Integration and analysis of multi-omics data give us a broad view of tumours, which can improve clinical decision making. Omics data, mainly DNA methylation and gene expression profiles are usually high dimensional data with a lot of molecular features. In recent years, variational autoencoders (VAE) have been extensively used in embedding image and text data into lower dimensional latent spaces. In our project, we extend the idea of using a VAE model for low dimensional latent space extraction with the self-supervised learning technique of feature subsetting. With VAEs, the key idea is to make the model learn meaningful representations from different types of omics data, which could then be used for downstream tasks such as cancer type classification. The main goals are to overcome the curse of dimensionality and integrate methylation and expression data to combine information about different aspects of same tissue samples, and hopefully extract biologically relevant features. Our extension involves training encoder and decoder to reconstruct the data from just a subset of it. By doing this, we force the model to encode most important information in the latent representation. We also added an identity to the subsets so that the model knows which subset is being fed into it during training and testing. We experimented with our approach and found that SubOmiEmbed produces comparable results to the baseline OmiEmbed with a much smaller network and by using just a subset of the data. This work can be improved to integrate mutation-based genomic data as well.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge