Self-omics: A Self-supervised Learning Framework for Multi-omics Cancer Data
Paper and Code
Oct 03, 2022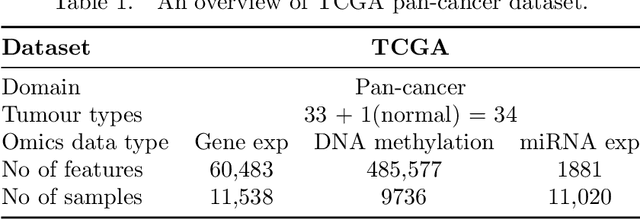
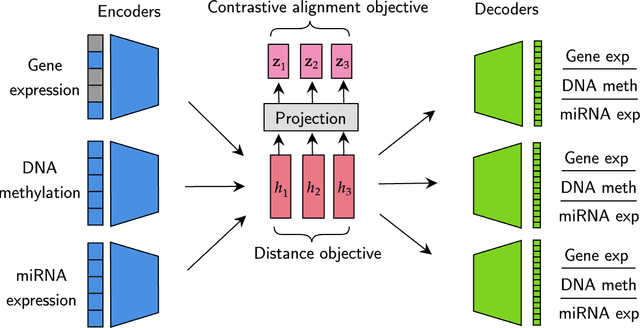
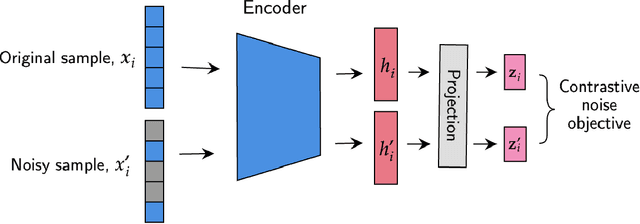
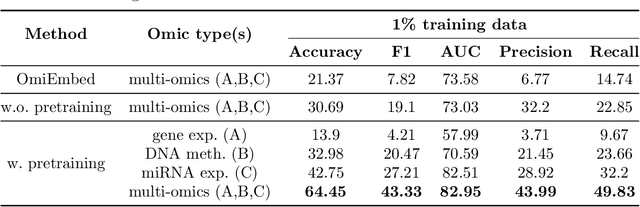
We have gained access to vast amounts of multi-omics data thanks to Next Generation Sequencing. However, it is challenging to analyse this data due to its high dimensionality and much of it not being annotated. Lack of annotated data is a significant problem in machine learning, and Self-Supervised Learning (SSL) methods are typically used to deal with limited labelled data. However, there is a lack of studies that use SSL methods to exploit inter-omics relationships on unlabelled multi-omics data. In this work, we develop a novel and efficient pre-training paradigm that consists of various SSL components, including but not limited to contrastive alignment, data recovery from corrupted samples, and using one type of omics data to recover other omic types. Our pre-training paradigm improves performance on downstream tasks with limited labelled data. We show that our approach outperforms the state-of-the-art method in cancer type classification on the TCGA pan-cancer dataset in semi-supervised setting. Moreover, we show that the encoders that are pre-trained using our approach can be used as powerful feature extractors even without fine-tuning. Our ablation study shows that the method is not overly dependent on any pretext task component. The network architectures in our approach are designed to handle missing omic types and multiple datasets for pre-training and downstream training. Our pre-training paradigm can be extended to perform zero-shot classification of rare cancers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge