Sascha E. A. Muenzing
Analyzing Regional Organization of the Human Hippocampus in 3D-PLI Using Contrastive Learning and Geometric Unfolding
Feb 27, 2024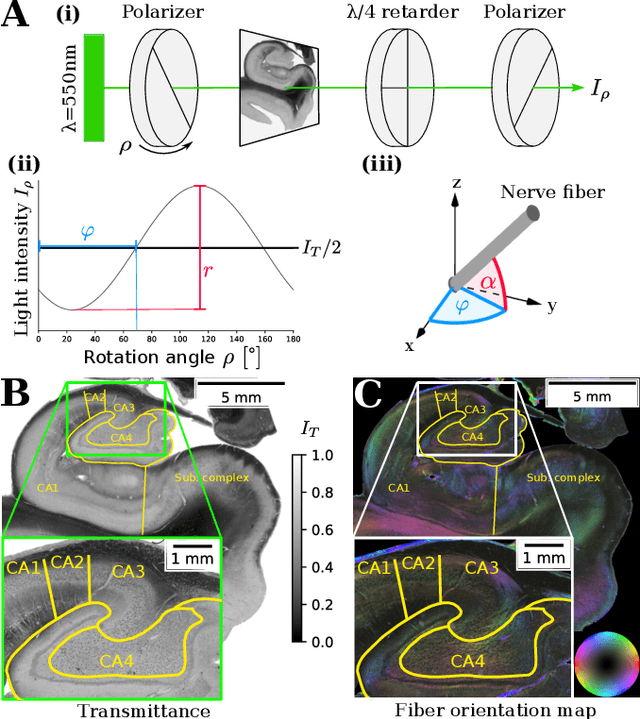
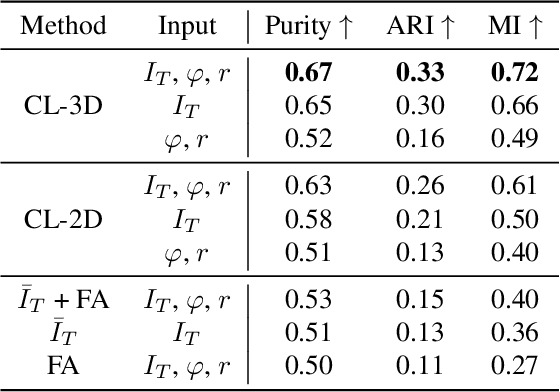
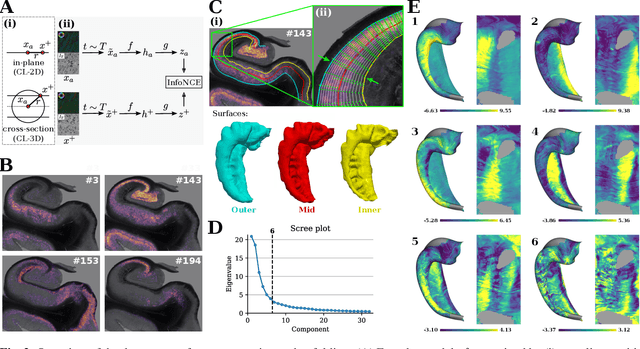
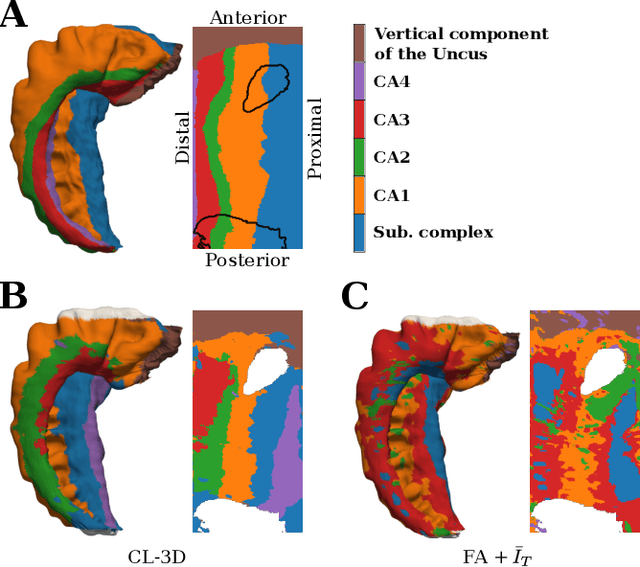
Abstract:Understanding the cortical organization of the human brain requires interpretable descriptors for distinct structural and functional imaging data. 3D polarized light imaging (3D-PLI) is an imaging modality for visualizing fiber architecture in postmortem brains with high resolution that also captures the presence of cell bodies, for example, to identify hippocampal subfields. The rich texture in 3D-PLI images, however, makes this modality particularly difficult to analyze and best practices for characterizing architectonic patterns still need to be established. In this work, we demonstrate a novel method to analyze the regional organization of the human hippocampus in 3D-PLI by combining recent advances in unfolding methods with deep texture features obtained using a self-supervised contrastive learning approach. We identify clusters in the representations that correspond well with classical descriptions of hippocampal subfields, lending validity to the developed methodology.
Self-Supervised Representation Learning for Nerve Fiber Distribution Patterns in 3D-PLI
Jan 30, 2024Abstract:A comprehensive understanding of the organizational principles in the human brain requires, among other factors, well-quantifiable descriptors of nerve fiber architecture. Three-dimensional polarized light imaging (3D-PLI) is a microscopic imaging technique that enables insights into the fine-grained organization of myelinated nerve fibers with high resolution. Descriptors characterizing the fiber architecture observed in 3D-PLI would enable downstream analysis tasks such as multimodal correlation studies, clustering, and mapping. However, best practices for observer-independent characterization of fiber architecture in 3D-PLI are not yet available. To this end, we propose the application of a fully data-driven approach to characterize nerve fiber architecture in 3D-PLI images using self-supervised representation learning. We introduce a 3D-Context Contrastive Learning (CL-3D) objective that utilizes the spatial neighborhood of texture examples across histological brain sections of a 3D reconstructed volume to sample positive pairs for contrastive learning. We combine this sampling strategy with specifically designed image augmentations to gain robustness to typical variations in 3D-PLI parameter maps. The approach is demonstrated for the 3D reconstructed occipital lobe of a vervet monkey brain. We show that extracted features are highly sensitive to different configurations of nerve fibers, yet robust to variations between consecutive brain sections arising from histological processing. We demonstrate their practical applicability for retrieving clusters of homogeneous fiber architecture and performing data mining for interactively selected templates of specific components of fiber architecture such as U-fibers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge