Sarah Zhao
Scalable Amortized GPLVMs for Single Cell Transcriptomics Data
May 06, 2024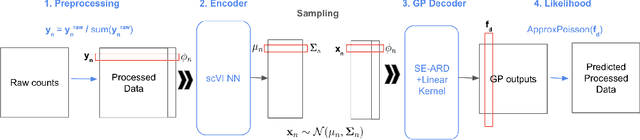
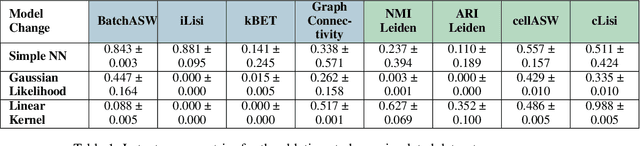
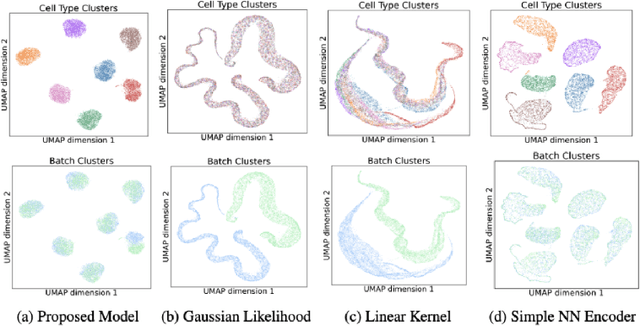
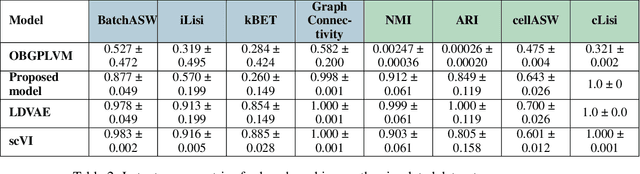
Abstract:Dimensionality reduction is crucial for analyzing large-scale single-cell RNA-seq data. Gaussian Process Latent Variable Models (GPLVMs) offer an interpretable dimensionality reduction method, but current scalable models lack effectiveness in clustering cell types. We introduce an improved model, the amortized stochastic variational Bayesian GPLVM (BGPLVM), tailored for single-cell RNA-seq with specialized encoder, kernel, and likelihood designs. This model matches the performance of the leading single-cell variational inference (scVI) approach on synthetic and real-world COVID datasets and effectively incorporates cell-cycle and batch information to reveal more interpretable latent structures as we demonstrate on an innate immunity dataset.
Geodesic Mode Connectivity
Aug 24, 2023Abstract:Mode connectivity is a phenomenon where trained models are connected by a path of low loss. We reframe this in the context of Information Geometry, where neural networks are studied as spaces of parameterized distributions with curved geometry. We hypothesize that shortest paths in these spaces, known as geodesics, correspond to mode-connecting paths in the loss landscape. We propose an algorithm to approximate geodesics and demonstrate that they achieve mode connectivity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge