Saqib Qamar
SAMA-UNet: Enhancing Medical Image Segmentation with Self-Adaptive Mamba-Like Attention and Causal-Resonance Learning
May 21, 2025Abstract:Medical image segmentation plays an important role in various clinical applications, but existing models often struggle with the computational inefficiencies and challenges posed by complex medical data. State Space Sequence Models (SSMs) have demonstrated promise in modeling long-range dependencies with linear computational complexity, yet their application in medical image segmentation remains hindered by incompatibilities with image tokens and autoregressive assumptions. Moreover, it is difficult to achieve a balance in capturing both local fine-grained information and global semantic dependencies. To address these challenges, we introduce SAMA-UNet, a novel architecture for medical image segmentation. A key innovation is the Self-Adaptive Mamba-like Aggregated Attention (SAMA) block, which integrates contextual self-attention with dynamic weight modulation to prioritise the most relevant features based on local and global contexts. This approach reduces computational complexity and improves the representation of complex image features across multiple scales. We also suggest the Causal-Resonance Multi-Scale Module (CR-MSM), which enhances the flow of information between the encoder and decoder by using causal resonance learning. This mechanism allows the model to automatically adjust feature resolution and causal dependencies across scales, leading to better semantic alignment between the low-level and high-level features in U-shaped architectures. Experiments on MRI, CT, and endoscopy images show that SAMA-UNet performs better in segmentation accuracy than current methods using CNN, Transformer, and Mamba. The implementation is publicly available at GitHub.
ScaleFusionNet: Transformer-Guided Multi-Scale Feature Fusion for Skin Lesion Segmentation
Mar 05, 2025Abstract:Melanoma is a malignant tumor originating from skin cell lesions. Accurate and efficient segmentation of skin lesions is essential for quantitative medical analysis but remains challenging. To address this, we propose ScaleFusionNet, a segmentation model that integrates Cross-Attention Transformer Module (CATM) and AdaptiveFusionBlock to enhance feature extraction and fusion. The model employs a hybrid architecture encoder that effectively captures both local and global features. We introduce CATM, which utilizes Swin Transformer Blocks and Cross Attention Fusion (CAF) to adaptively refine encoder-decoder feature fusion, reducing semantic gaps and improving segmentation accuracy. Additionally, the AdaptiveFusionBlock is improved by integrating adaptive multi-scale fusion, where Swin Transformer-based attention complements deformable convolution-based multi-scale feature extraction. This enhancement refines lesion boundaries and preserves fine-grained details. ScaleFusionNet achieves Dice scores of 92.94% and 91.65% on ISIC-2016 and ISIC-2018 datasets, respectively, demonstrating its effectiveness in skin lesion analysis. Our code implementation is publicly available at GitHub.
Segmentation and Characterization of Macerated Fibers and Vessels Using Deep Learning
Jan 30, 2024Abstract:Purpose: Wood comprises different cell types, such as fibers and vessels, defining its properties. Studying their shape, size, and arrangement in microscopic images is crucial for understanding wood samples. Typically, this involves macerating (soaking) samples in a solution to separate cells, then spreading them on slides for imaging with a microscope that covers a wide area, capturing thousands of cells. However, these cells often cluster and overlap in images, making the segmentation difficult and time-consuming using standard image-processing methods. Results: In this work, we develop an automatic deep learning segmentation approach that utilizes the one-stage YOLOv8 model for fast and accurate fiber and vessel segmentation and characterization in microscopy images. The model can analyze 32640 x 25920 pixels images and demonstrate effective cell detection and segmentation, achieving a mAP_0.5-0.95 of 78 %. To assess the model's robustness, we examined fibers from a genetically modified tree line known for longer fibers. The outcomes were comparable to previous manual measurements. Additionally, we created a user-friendly web application for image analysis and provided the code for use on Google Colab. Conclusion: By leveraging YOLOv8's advances, this work provides a deep learning solution to enable efficient quantification and analysis of wood cells suitable for practical applications.
HI-Net: Hyperdense Inception 3D UNet for Brain Tumor Segmentation
Dec 12, 2020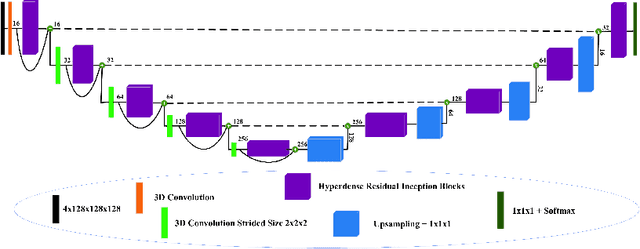
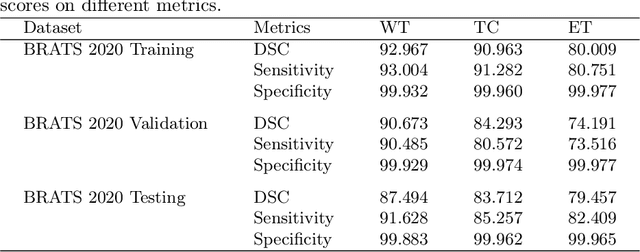
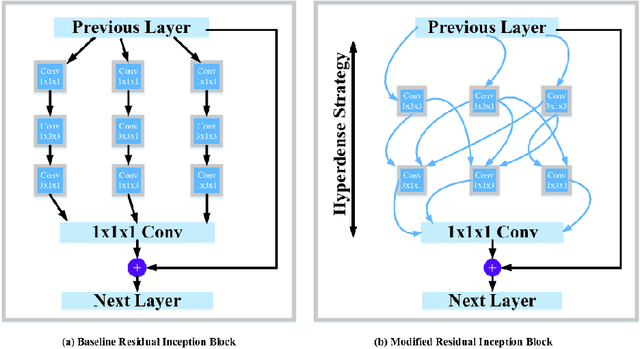
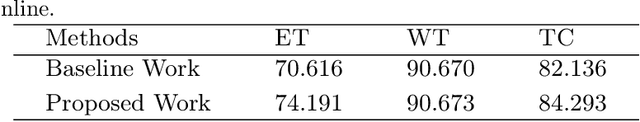
Abstract:The brain tumor segmentation task aims to classify tissue into the whole tumor (WT), tumor core (TC), and enhancing tumor (ET) classes using multimodel MRI images. Quantitative analysis of brain tumors is critical for clinical decision making. While manual segmentation is tedious, time-consuming, and subjective, this task is at the same time very challenging to automatic segmentation methods. Thanks to the powerful learning ability, convolutional neural networks (CNNs), mainly fully convolutional networks, have shown promising brain tumor segmentation. This paper further boosts the performance of brain tumor segmentation by proposing hyperdense inception 3D UNet (HI-Net), which captures multi-scale information by stacking factorization of 3D weighted convolutional layers in the residual inception block. We use hyper dense connections among factorized convolutional layers to extract more contexual information, with the help of features reusability. We use a dice loss function to cope with class imbalances. We validate the proposed architecture on the multi-modal brain tumor segmentation challenges (BRATS) 2020 testing dataset. Preliminary results on the BRATS 2020 testing set show that achieved by our proposed approach, the dice (DSC) scores of ET, WT, and TC are 0.79457, 0.87494, and 0.83712, respectively.
Context Aware 3D UNet for Brain Tumor Segmentation
Oct 25, 2020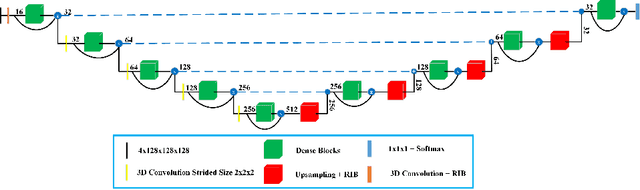
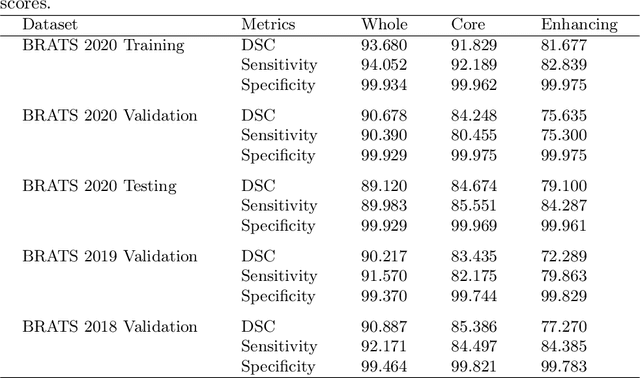
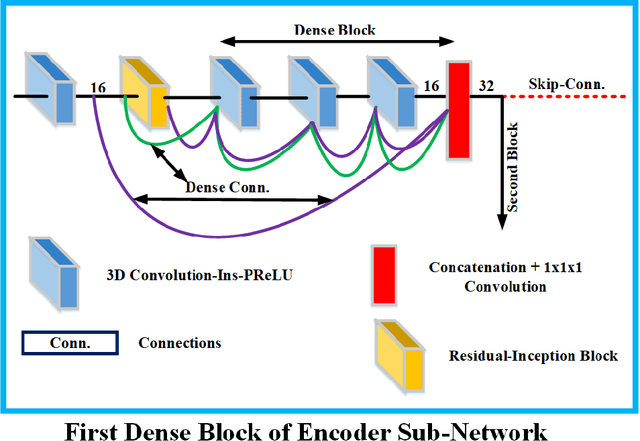
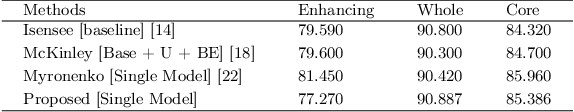
Abstract:Deep convolutional neural network (CNN) achieves remarkable performance for medical image analysis. UNet is the primary source in the performance of 3D CNN architectures for medical imaging tasks, including brain tumor segmentation. The skip connection in the UNet architecture concatenates features from both encoder and decoder paths to extract multi-contexual information from image data. The multi-scaled features play an essential role in brain tumor segmentation. However, the limited use of features can degrade the performance of the UNet approach for segmentation. In this paper, we propose a modified UNet architecture for brain tumor segmentation. In the proposed architecture, we used densely connected blocks in both encoder and decoder paths to extract multi-contexual information from the concept of feature reusability. The proposed residual inception blocks (RIB) are used to extract local and global information by merging features of different kernel sizes. We validate the proposed architecture on the multimodal brain tumor segmentation challenges (BRATS) 2020 testing dataset. The dice (DSC) scores of the whole tumor (WT), tumor core (TC), and enhancement tumor (ET) are 89.12%, 84.74%, and 79.12%, respectively. Our proposed work is in the top ten methods based on the dice scores of the testing dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge