Context Aware 3D UNet for Brain Tumor Segmentation
Paper and Code
Oct 25, 2020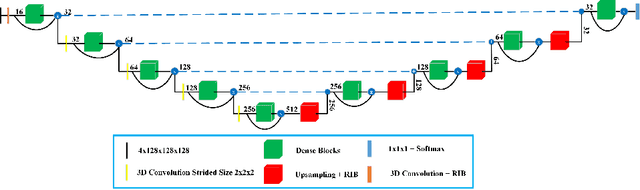
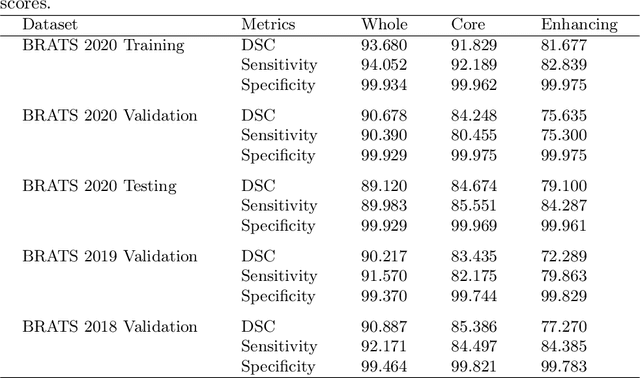
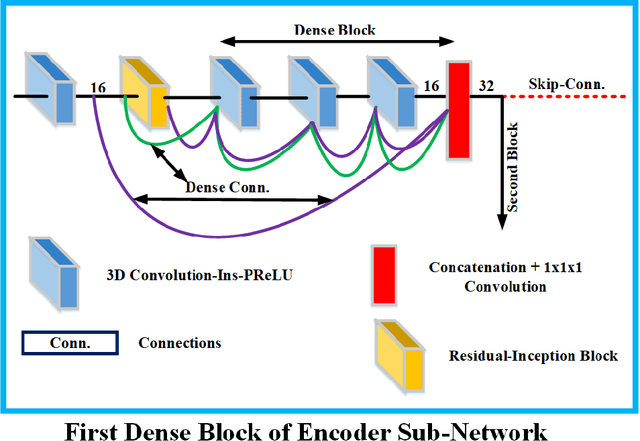
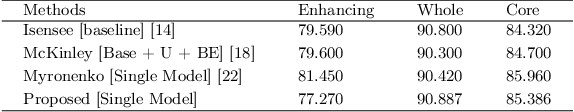
Deep convolutional neural network (CNN) achieves remarkable performance for medical image analysis. UNet is the primary source in the performance of 3D CNN architectures for medical imaging tasks, including brain tumor segmentation. The skip connection in the UNet architecture concatenates features from both encoder and decoder paths to extract multi-contexual information from image data. The multi-scaled features play an essential role in brain tumor segmentation. However, the limited use of features can degrade the performance of the UNet approach for segmentation. In this paper, we propose a modified UNet architecture for brain tumor segmentation. In the proposed architecture, we used densely connected blocks in both encoder and decoder paths to extract multi-contexual information from the concept of feature reusability. The proposed residual inception blocks (RIB) are used to extract local and global information by merging features of different kernel sizes. We validate the proposed architecture on the multimodal brain tumor segmentation challenges (BRATS) 2020 testing dataset. The dice (DSC) scores of the whole tumor (WT), tumor core (TC), and enhancement tumor (ET) are 89.12%, 84.74%, and 79.12%, respectively. Our proposed work is in the top ten methods based on the dice scores of the testing dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge