Sanmitra Ghosh
Light-Weight Diffusion Multiplier and Uncertainty Quantification for Fourier Neural Operators
Aug 01, 2025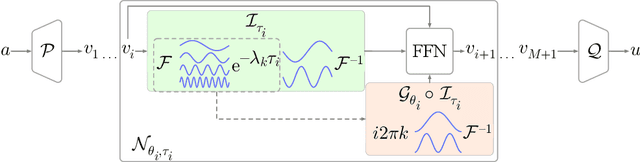
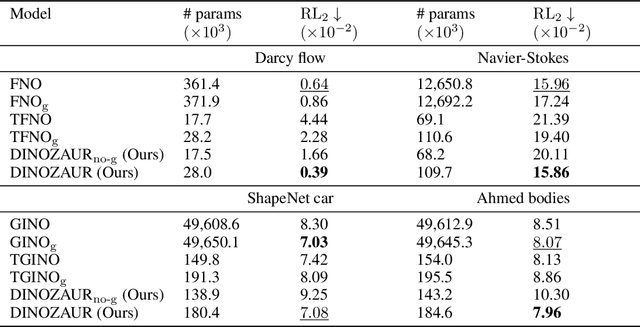
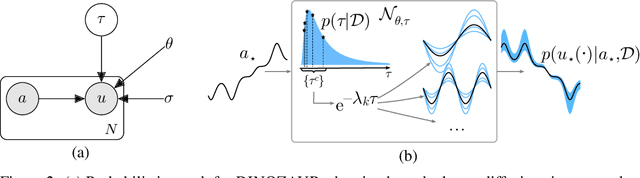
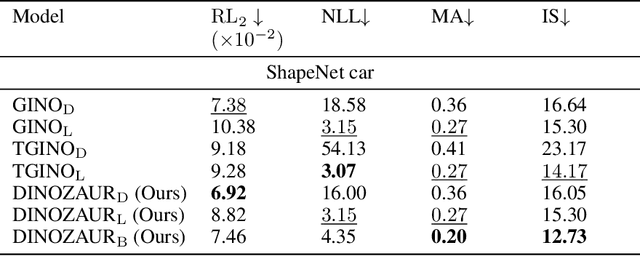
Abstract:Operator learning is a powerful paradigm for solving partial differential equations, with Fourier Neural Operators serving as a widely adopted foundation. However, FNOs face significant scalability challenges due to overparameterization and offer no native uncertainty quantification -- a key requirement for reliable scientific and engineering applications. Instead, neural operators rely on post hoc UQ methods that ignore geometric inductive biases. In this work, we introduce DINOZAUR: a diffusion-based neural operator parametrization with uncertainty quantification. Inspired by the structure of the heat kernel, DINOZAUR replaces the dense tensor multiplier in FNOs with a dimensionality-independent diffusion multiplier that has a single learnable time parameter per channel, drastically reducing parameter count and memory footprint without compromising predictive performance. By defining priors over those time parameters, we cast DINOZAUR as a Bayesian neural operator to yield spatially correlated outputs and calibrated uncertainty estimates. Our method achieves competitive or superior performance across several PDE benchmarks while providing efficient uncertainty quantification.
Sample-efficient neural likelihood-free Bayesian inference of implicit HMMs
May 02, 2024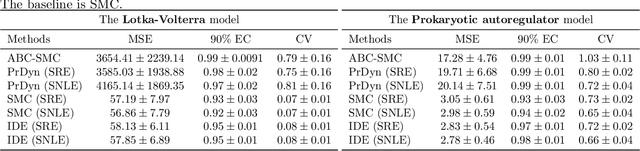
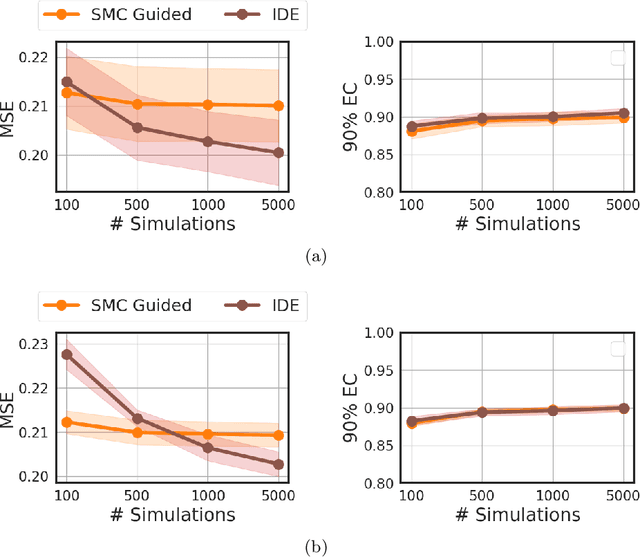

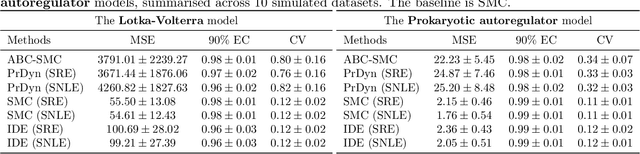
Abstract:Likelihood-free inference methods based on neural conditional density estimation were shown to drastically reduce the simulation burden in comparison to classical methods such as ABC. When applied in the context of any latent variable model, such as a Hidden Markov model (HMM), these methods are designed to only estimate the parameters, rather than the joint distribution of the parameters and the hidden states. Naive application of these methods to a HMM, ignoring the inference of this joint posterior distribution, will thus produce an inaccurate estimate of the posterior predictive distribution, in turn hampering the assessment of goodness-of-fit. To rectify this problem, we propose a novel, sample-efficient likelihood-free method for estimating the high-dimensional hidden states of an implicit HMM. Our approach relies on learning directly the intractable posterior distribution of the hidden states, using an autoregressive-flow, by exploiting the Markov property. Upon evaluating our approach on some implicit HMMs, we found that the quality of the estimates retrieved using our method is comparable to what can be achieved using a much more computationally expensive SMC algorithm.
Considering discrepancy when calibrating a mechanistic electrophysiology model
Jan 13, 2020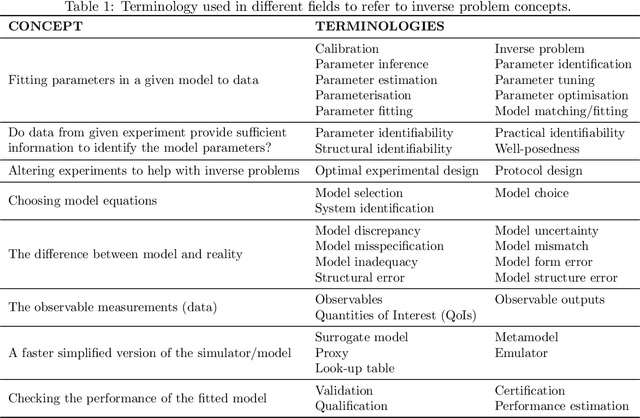
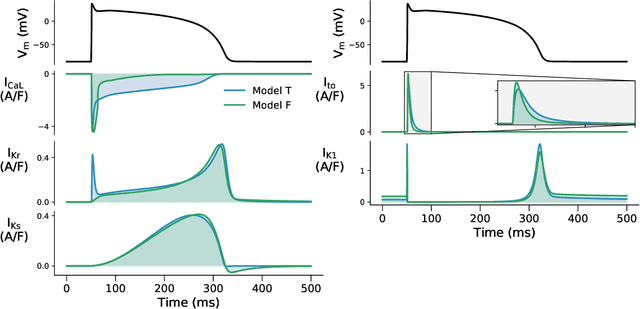
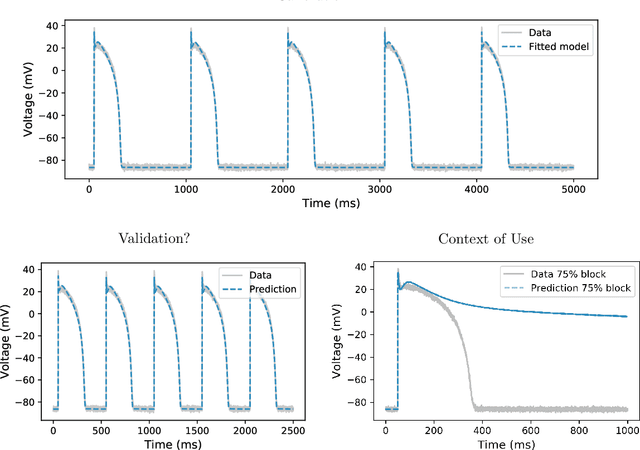
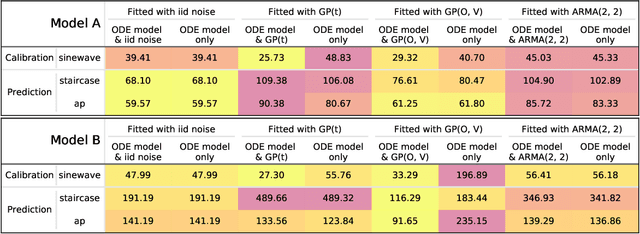
Abstract:Uncertainty quantification (UQ) is a vital step in using mathematical models and simulations to take decisions. The field of cardiac simulation has begun to explore and adopt UQ methods to characterise uncertainty in model inputs and how that propagates through to outputs or predictions. In this perspective piece we draw attention to an important and under-addressed source of uncertainty in our predictions --- that of uncertainty in the model structure or the equations themselves. The difference between imperfect models and reality is termed model discrepancy, and we are often uncertain as to the size and consequences of this discrepancy. Here we provide two examples of the consequences of discrepancy when calibrating models at the ion channel and action potential scales. Furthermore, we attempt to account for this discrepancy when calibrating and validating an ion channel model using different methods, based on modelling the discrepancy using Gaussian processes (GPs) and autoregressive-moving-average (ARMA) models, then highlight the advantages and shortcomings of each approach. Finally, suggestions and lines of enquiry for future work are provided.
Fast Approximate Bayesian Computation for Estimating Parameters in Differential Equations
Jul 17, 2015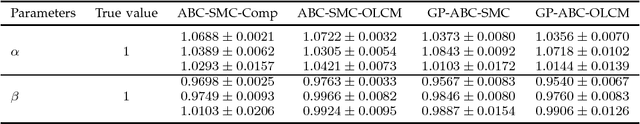
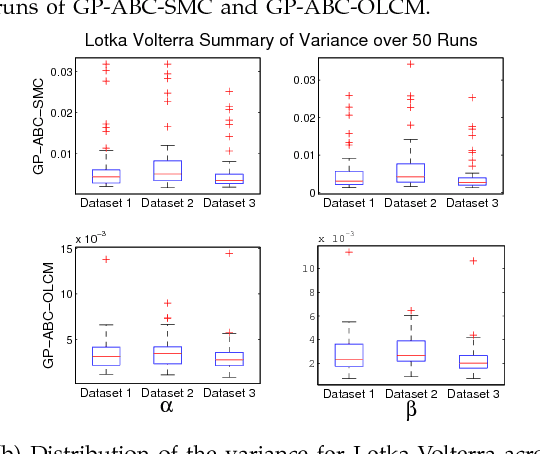
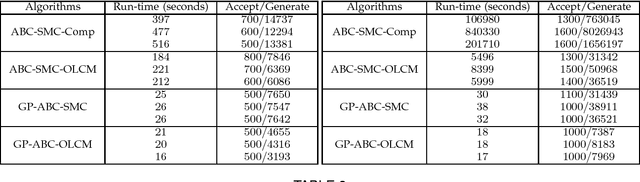
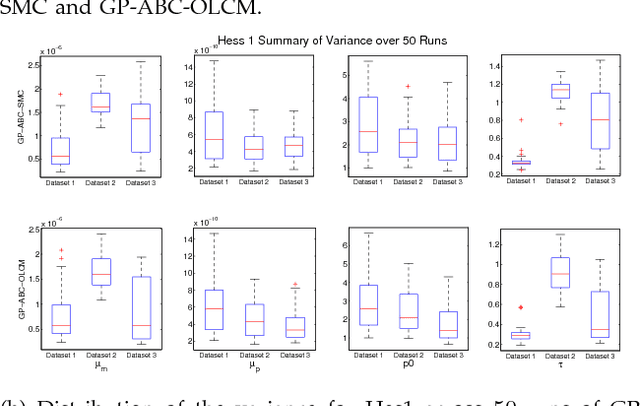
Abstract:Approximate Bayesian computation (ABC) using a sequential Monte Carlo method provides a comprehensive platform for parameter estimation, model selection and sensitivity analysis in differential equations. However, this method, like other Monte Carlo methods, incurs a significant computational cost as it requires explicit numerical integration of differential equations to carry out inference. In this paper we propose a novel method for circumventing the requirement of explicit integration by using derivatives of Gaussian processes to smooth the observations from which parameters are estimated. We evaluate our methods using synthetic data generated from model biological systems described by ordinary and delay differential equations. Upon comparing the performance of our method to existing ABC techniques, we demonstrate that it produces comparably reliable parameter estimates at a significantly reduced execution time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge