Sanjay Kalra
SF2Former: Amyotrophic Lateral Sclerosis Identification From Multi-center MRI Data Using Spatial and Frequency Fusion Transformer
Feb 28, 2023



Abstract:Amyotrophic Lateral Sclerosis (ALS) is a complex neurodegenerative disorder involving motor neuron degeneration. Significant research has begun to establish brain magnetic resonance imaging (MRI) as a potential biomarker to diagnose and monitor the state of the disease. Deep learning has turned into a prominent class of machine learning programs in computer vision and has been successfully employed to solve diverse medical image analysis tasks. However, deep learning-based methods applied to neuroimaging have not achieved superior performance in ALS patients classification from healthy controls due to having insignificant structural changes correlated with pathological features. Therefore, the critical challenge in deep models is to determine useful discriminative features with limited training data. By exploiting the long-range relationship of image features, this study introduces a framework named SF2Former that leverages vision transformer architecture's power to distinguish the ALS subjects from the control group. To further improve the network's performance, spatial and frequency domain information are combined because MRI scans are captured in the frequency domain before being converted to the spatial domain. The proposed framework is trained with a set of consecutive coronal 2D slices, which uses the pre-trained weights on ImageNet by leveraging transfer learning. Finally, a majority voting scheme has been employed to those coronal slices of a particular subject to produce the final classification decision. Our proposed architecture has been thoroughly assessed with multi-modal neuroimaging data using two well-organized versions of the Canadian ALS Neuroimaging Consortium (CALSNIC) multi-center datasets. The experimental results demonstrate the superiority of our proposed strategy in terms of classification accuracy compared with several popular deep learning-based techniques.
Texture Classification of MR Images of the Brain in ALS using CoHOG
Sep 25, 2017
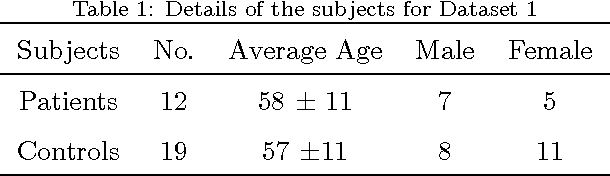
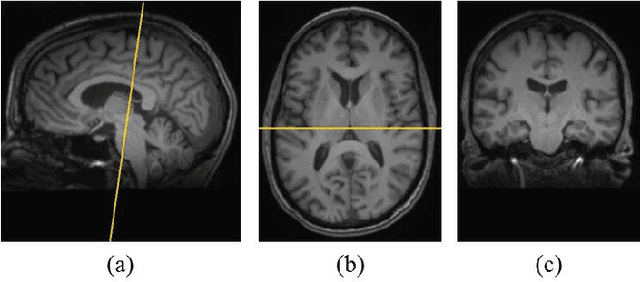
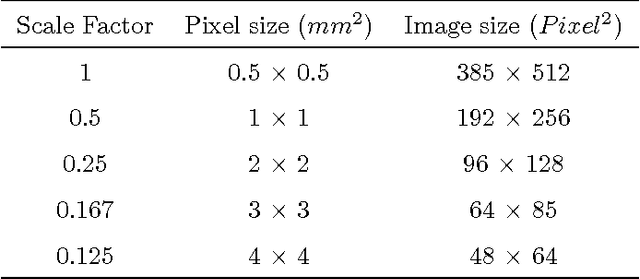
Abstract:Texture analysis is a well-known research topic in computer vision and image processing and has many applications. Gradient-based texture methods have become popular in classification problems. For the first time we extend a well-known gradient-based method, Co-occurrence Histograms of Oriented Gradients (CoHOG) to extract texture features from 2D Magnetic Resonance Images (MRI). Unlike the original CoHOG method, we use the whole image instead of sub-regions for feature calculation. Also, we use a larger neighborhood size. Gradient orientations of the image pixels are calculated using Sobel, Gaussian Derivative (GD) and Local Frequency Descriptor Gradient (LFDG) operators. The extracted feature vector size is very large and classification using a large number of similar features does not provide the best results. In our proposed method, for the first time to our best knowledge, only a minimum number of significant features are selected using area under the receiver operator characteristic (ROC) curve (AUC) thresholds with <= 0.01. In this paper, we apply the proposed method to classify Amyotrophic Lateral Sclerosis (ALS) patients from the controls. It is observed that selected texture features from downsampled images are significantly different between patients and controls. These features are used in a linear support vector machine (SVM) classifier to determine the classification accuracy. Optimal sensitivity and specificity are also calculated. Three different cohort datasets are used in the experiments. The performance of the proposed method using three gradient operators and two different neighborhood sizes is analyzed. Region based analysis is performed to demonstrate that significant changes between patients and controls are limited to the motor cortex.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge