Sandhya Prabhakaran
A Nonparametric Multi-view Model for Estimating Cell Type-Specific Gene Regulatory Networks
Feb 21, 2019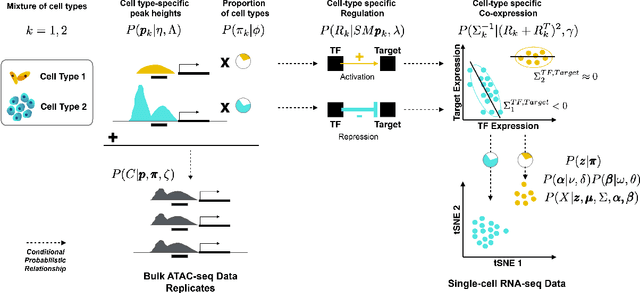
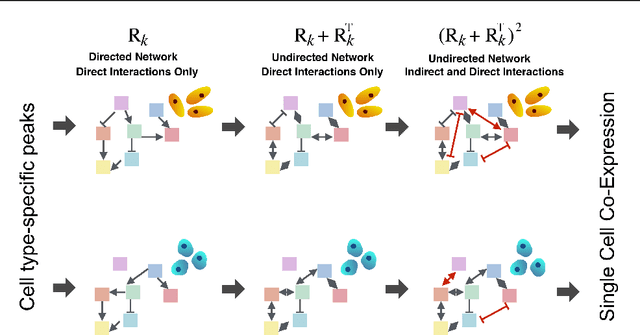


Abstract:We present a Bayesian hierarchical multi-view mixture model termed Symphony that simultaneously learns clusters of cells representing cell types and their underlying gene regulatory networks by integrating data from two views: single-cell gene expression data and paired epigenetic data, which is informative of gene-gene interactions. This model improves interpretation of clusters as cell types with similar expression patterns as well as regulatory networks driving expression, by explaining gene-gene covariances with the biological machinery regulating gene expression. We show the theoretical advantages of the multi-view learning approach and present a Variational EM inference procedure. We demonstrate superior performance on both synthetic data and real genomic data with subtypes of peripheral blood cells compared to other methods.
Probabilistic Clustering of Time-Evolving Distance Data
Apr 14, 2015



Abstract:We present a novel probabilistic clustering model for objects that are represented via pairwise distances and observed at different time points. The proposed method utilizes the information given by adjacent time points to find the underlying cluster structure and obtain a smooth cluster evolution. This approach allows the number of objects and clusters to differ at every time point, and no identification on the identities of the objects is needed. Further, the model does not require the number of clusters being specified in advance -- they are instead determined automatically using a Dirichlet process prior. We validate our model on synthetic data showing that the proposed method is more accurate than state-of-the-art clustering methods. Finally, we use our dynamic clustering model to analyze and illustrate the evolution of brain cancer patients over time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge