Samuel St-Jean
Harmonization of diffusion MRI datasets with adaptive dictionary learning
Oct 30, 2019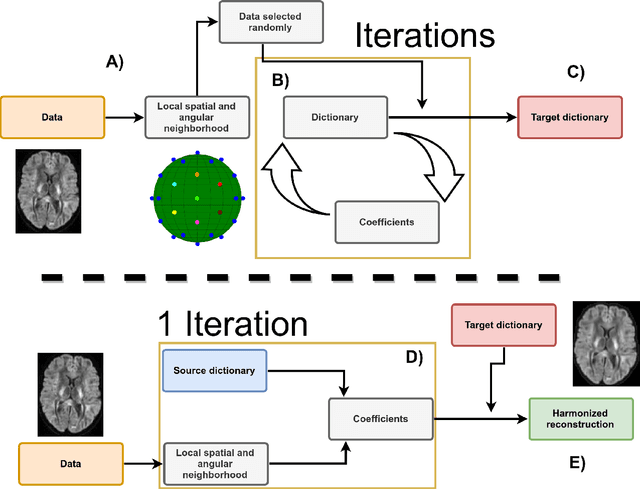
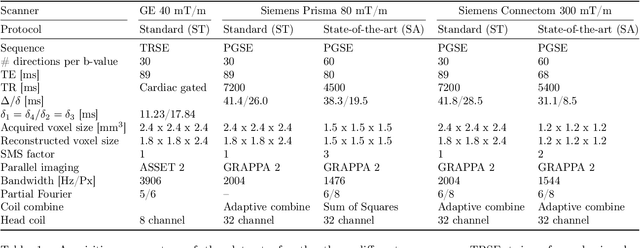
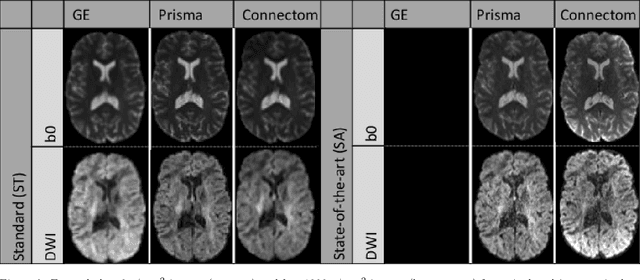
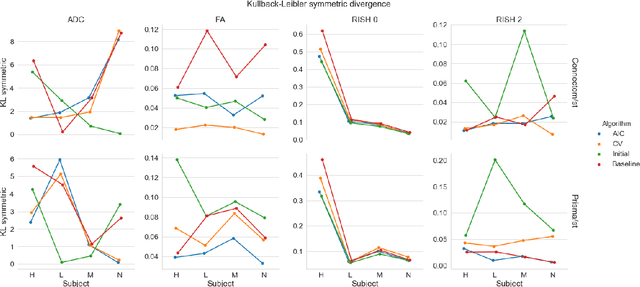
Abstract:Diffusion magnetic resonance imaging is a noninvasive imaging technique which can indirectly infer the microstructure of tissues and provide metrics which are subject to normal variability across subjects. Potentially abnormal values or features may yield essential information to support analysis of controls and patients cohorts, but subtle confounds affecting diffusion MRI, such as those due to difference in scanning protocols or hardware, can lead to systematic errors which could be mistaken for purely biologically driven variations amongst subjects. In this work, we propose a new harmonization algorithm based on adaptive dictionary learning to mitigate the unwanted variability caused by different scanner hardware while preserving the natural biological variability present in the data. Overcomplete dictionaries, which are learned automatically from the data and do not require paired samples, are then used to reconstruct the data from a different scanner, removing variability present in the source scanner in the process. We use the publicly available database from an international challenge to evaluate the method, which was acquired on three different scanners and with two different protocols, and propose a new mapping towards a scanner-agnostic space. Results show that the effect size of the four studied diffusion metrics is preserved while removing variability attributable to the scanner. Experiments with alterations using a free water compartment, which is not simulated in the training data, shows that the effect size induced by the alterations is also preserved after harmonization. The algorithm is freely available and could help multicenter studies in pooling their data, while removing scanner specific confounds, and increase statistical power in the process.
Automatic, fast and robust characterization of noise distributions for diffusion MRI
Oct 02, 2018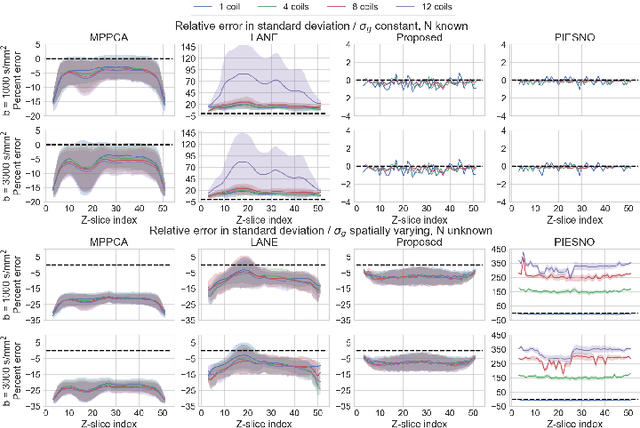
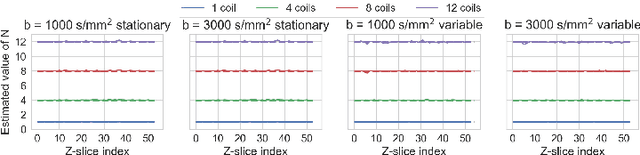
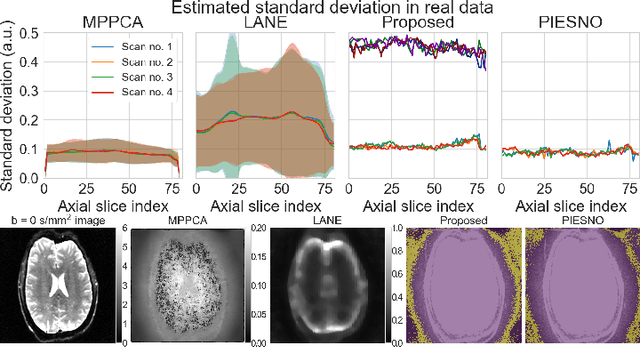
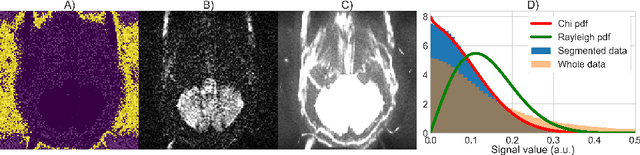
Abstract:Knowledge of the noise distribution in magnitude diffusion MRI images is the centerpiece to quantify uncertainties arising from the acquisition process. The use of parallel imaging methods, the number of receiver coils and imaging filters applied by the scanner, amongst other factors, dictate the resulting signal distribution. Accurate estimation beyond textbook Rician or noncentral chi distributions often requires information about the acquisition process (e.g. coils sensitivity maps or reconstruction coefficients), which is not usually available. We introduce a new method where a change of variable naturally gives rise to a particular form of the gamma distribution for background signals. The first moments and maximum likelihood estimators of this gamma distribution explicitly depend on the number of coils, making it possible to estimate all unknown parameters using only the magnitude data. A rejection step is used to make the method automatic and robust to artifacts. Experiments on synthetic datasets show that the proposed method can reliably estimate both the degrees of freedom and the standard deviation. The worst case errors range from below 2% (spatially uniform noise) to approximately 10% (spatially variable noise). Repeated acquisitions of in vivo datasets show that the estimated parameters are stable and have lower variances than compared methods.
* v2: added publisher DOI statement, fixed text typo in appendix A2
Non Local Spatial and Angular Matching : Enabling higher spatial resolution diffusion MRI datasets through adaptive denoising
Jun 23, 2016



Abstract:Diffusion magnetic resonance imaging datasets suffer from low Signal-to-Noise Ratio, especially at high b-values. Acquiring data at high b-values contains relevant information and is now of great interest for microstructural and connectomics studies. High noise levels bias the measurements due to the non-Gaussian nature of the noise, which in turn can lead to a false and biased estimation of the diffusion parameters. Additionally, the usage of in-plane acceleration techniques during the acquisition leads to a spatially varying noise distribution, which depends on the parallel acceleration method implemented on the scanner. This paper proposes a novel diffusion MRI denoising technique that can be used on all existing data, without adding to the scanning time. We first apply a statistical framework to convert the noise to Gaussian distributed noise, effectively removing the bias. We then introduce a spatially and angular adaptive denoising technique, the Non Local Spatial and Angular Matching (NLSAM) algorithm. Each volume is first decomposed in small 4D overlapping patches to capture the structure of the diffusion data and a dictionary of atoms is learned on those patches. A local sparse decomposition is then found by bounding the reconstruction error with the local noise variance. We compare against three other state-of-the-art denoising methods and show quantitative local and connectivity results on a synthetic phantom and on an in-vivo high resolution dataset. Overall, our method restores perceptual information, removes the noise bias in common diffusion metrics, restores the extracted peaks coherence and improves reproducibility of tractography. Our work paves the way for higher spatial resolution acquisition of diffusion MRI datasets, which could in turn reveal new anatomical details that are not discernible at the spatial resolution currently used by the diffusion MRI community.
* Code available : https://github.com/samuelstjean/nlsam Datasets available : https://github.com/samuelstjean/nlsam_data, Medical Image Analysis, 2016
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge