Harmonization of diffusion MRI datasets with adaptive dictionary learning
Paper and Code
Oct 30, 2019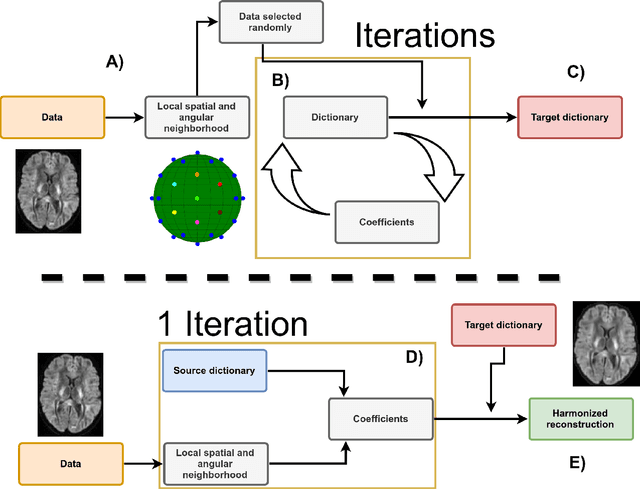
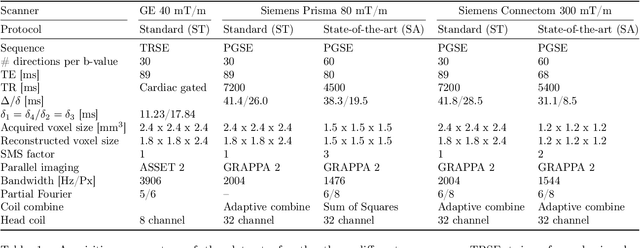
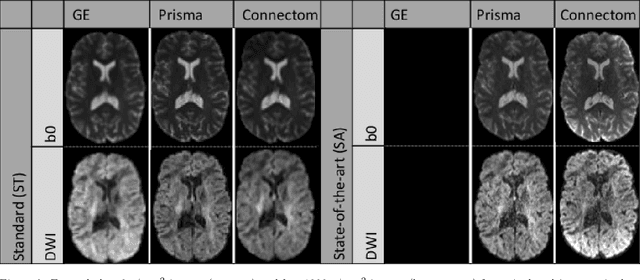
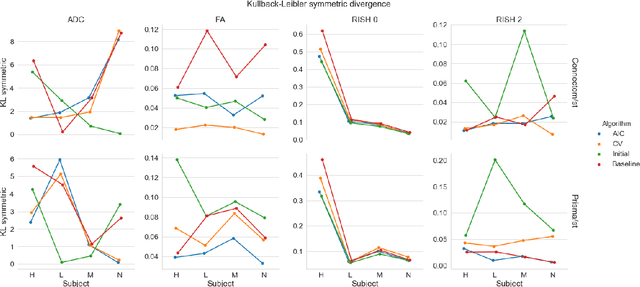
Diffusion magnetic resonance imaging is a noninvasive imaging technique which can indirectly infer the microstructure of tissues and provide metrics which are subject to normal variability across subjects. Potentially abnormal values or features may yield essential information to support analysis of controls and patients cohorts, but subtle confounds affecting diffusion MRI, such as those due to difference in scanning protocols or hardware, can lead to systematic errors which could be mistaken for purely biologically driven variations amongst subjects. In this work, we propose a new harmonization algorithm based on adaptive dictionary learning to mitigate the unwanted variability caused by different scanner hardware while preserving the natural biological variability present in the data. Overcomplete dictionaries, which are learned automatically from the data and do not require paired samples, are then used to reconstruct the data from a different scanner, removing variability present in the source scanner in the process. We use the publicly available database from an international challenge to evaluate the method, which was acquired on three different scanners and with two different protocols, and propose a new mapping towards a scanner-agnostic space. Results show that the effect size of the four studied diffusion metrics is preserved while removing variability attributable to the scanner. Experiments with alterations using a free water compartment, which is not simulated in the training data, shows that the effect size induced by the alterations is also preserved after harmonization. The algorithm is freely available and could help multicenter studies in pooling their data, while removing scanner specific confounds, and increase statistical power in the process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge